Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism
- PMID: 16126851
- PMCID: PMC1203358
- DOI: 10.1104/pp.105.066399
Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism
Abstract
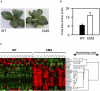
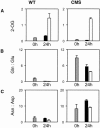
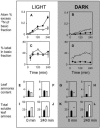
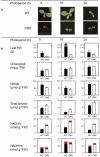
The Nicotiana sylvestris mutant, CMS, lacks the mitochondrial gene nad7 and functional complex I, and respires using low-affinity NADH (alternative) mitochondrial dehydrogenases. Here, we show that this adjustment of respiratory pathways is associated with a profound modification of foliar carbon-nitrogen balance. CMS leaves are characterized by abundant amino acids compared to either wild-type plants or CMS in which complex I function has been restored by nuclear transformation with the nad7 cDNA. The metabolite profile of CMS leaves is enriched in amino acids with low carbon/nitrogen and depleted in starch and 2-oxoglutarate. Deficiency in 2-oxoglutarate occurred despite increased citrate and malate and higher capacity of key anaplerotic enzymes, notably the mitochondrial NAD-dependent isocitrate dehydrogenase. The accumulation of nitrogen-rich amino acids was not accompanied by increased expression of enzymes involved in nitrogen assimilation. Partitioning of (15)N-nitrate into soluble amines was enhanced in CMS leaf discs compared to wild-type discs, especially in the dark. Analysis of pyridine nucleotides showed that both NAD and NADH were increased by 2-fold in CMS leaves. The growth retardation of CMS relative to the wild type was highly dependent on photoperiod, but at all photoperiod regimes the link between high contents of amino acids and NADH was observed. Together, the data provide strong evidence that (1) NADH availability is a critical factor in influencing the rate of nitrate assimilation and that (2) NAD status plays a crucial role in coordinating ammonia assimilation with the anaplerotic production of carbon skeletons.
Figures



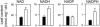

References
-
- Bai YD, Hajek P, Chomyn A, Chan E, Seo BB, Matsuno-Yagi A, Yagi T, Attardi G (2001) Lack of complex I activity in human cells carrying a mutation in MtDNA-encoded ND4 subunit is corrected by the Saccharomyces cerevisiae NADH-quinone oxidoreductase (NDI1) gene. J Biol Chem 276: 38808–38813 - PubMed
-
- Cousins AB, Bloom AJ (2004) Oxygen consumption during leaf nitrate assimilation in a C-3 and C-4 plant: the role of mitochondrial respiration. Plant Cell Environ 27: 1537–1545
-
- Douce R, Manella CA, Bonner WD (1973) The external NADH dehydrogenases of intact plant mitochondria. Biochim Biophys Acta 292: 105–116 - PubMed
-
- Dubois F, Brugiere N, Sangwan RS, Hirel B (1996) Localization of tobacco cytosolic glutamine synthetase enzymes and the corresponding transcripts shows organ- and cell-specific patterns of protein synthesis and gene expression. Plant Mol Biol 31: 803–817 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

