Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation
- PMID: 16131587
- PMCID: PMC1370832
- DOI: 10.1261/rna.2107305
Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation
Abstract
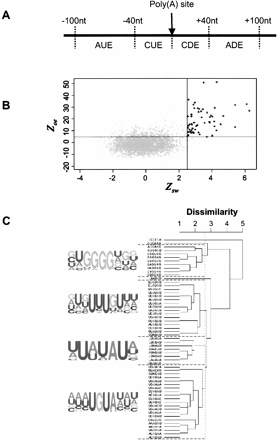
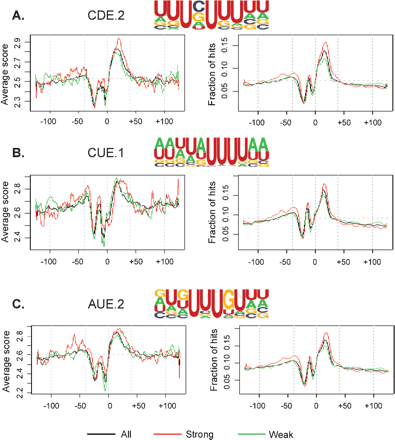
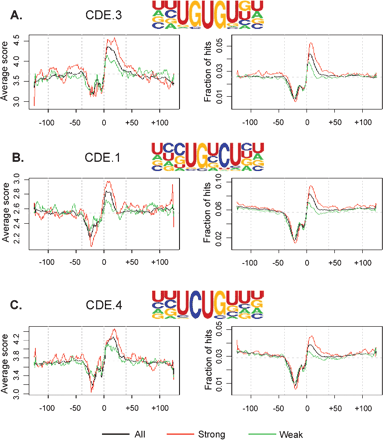
Polyadenylation is an essential step for the maturation of almost all cellular mRNAs in eukaryotes. In human cells, most poly(A) sites are flanked by the upstream AAUAAA hexamer or a close variant, and downstream U/GU-rich elements. In yeast and plants, additional cis elements have been found to be located upstream of the poly(A) site, including UGUA, UAUA, and U-rich elements. In this study, we have developed a computer program named PROBE (Polyadenylation-Related Oligonucleotide Bidimensional Enrichment) to identify cis elements that may play regulatory roles in mRNA polyadenylation. By comparing human genomic sequences surrounding frequently used poly(A) sites with those surrounding less frequently used ones, we found that cis elements occurring in yeast and plants also exist in human poly(A) regions, including the upstream U-rich elements, and UAUA and UGUA elements. In addition, several novel elements were found to be associated with human poly(A) sites, including several G-rich elements. Thus, we suggest that many cis elements are evolutionarily conserved among eukaryotes, and human poly(A) sites have an additional set of cis elements that may be involved in the regulation of mRNA polyadenylation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
