Organization of supercoil domains and their reorganization by transcription
- PMID: 16135220
- PMCID: PMC1382059
- DOI: 10.1111/j.1365-2958.2005.04796.x
Organization of supercoil domains and their reorganization by transcription
Abstract
During a normal cell cycle, chromosomes are exposed to many biochemical reactions that require specific types of DNA movement. Separation forces move replicated chromosomes into separate sister cell compartments during cell division, and the contemporaneous acts of DNA replication, RNA transcription and cotranscriptional translation of membrane proteins cause specific regions of DNA to twist, writhe and expand or contract. Recent experiments indicate that a dynamic and stochastic mechanism creates supercoil DNA domains soon after DNA replication. Domain structure is subsequently reorganized by RNA transcription. Examples of transcription-dependent chromosome remodelling are also emerging from eukaryotic cell systems.
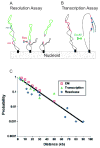
Figures

References
-
- Artsimovitch, I. (2005) Control of transcription termination and antitermination. In The Bacterial Chromosome Higgins, N.P. (ed.). Washington, DC: American Society for Microbiology Press, pp. 311–326.
-
- Aussel L, Barre FX, Aryoy M, Stasiak A, Stasiak AZ, Sherratt DJ. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. - PubMed
-
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

