Mutagenesis analysis of the rGTP-specific binding site of hepatitis C virus RNA-dependent RNA polymerase
- PMID: 16140738
- PMCID: PMC1212605
- DOI: 10.1128/JVI.79.18.11607-11617.2005
Mutagenesis analysis of the rGTP-specific binding site of hepatitis C virus RNA-dependent RNA polymerase
Abstract
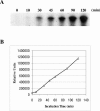
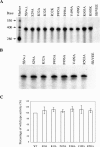
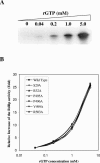
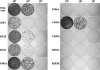
Hepatitis C virus (HCV) nonstructural protein 5B (NS5B) is the virus-encoded RNA-dependent RNA polymerase (RdRp) essential for HCV RNA replication. An earlier crystallographic study identified a rGTP-specific binding site lying at the surface between the thumb domain and the fingertip about 30 A away from the active site of the HCV RdRp (S. Bressanelli, L. Tomei, F. A. Rey, and R. De Francesco, J. Virol 76:3482-3492, 2002). To determine its physiological importance, we performed a systematic mutagenesis analysis of the rGTP-specific binding pocket by amino acid substitutions. Effects of mutations of the rGTP-specific binding site on enzymatic activity were determined by an in vitro RdRp assay, while effects of mutations on HCV RNA replication were examined by cell colony formation, as well as by transient replication of subgenomic HCV RNAs. Results derived from these studies demonstrate that amino acid substitutions of the rGTP-specific binding pocket did not significantly affect the in vitro RdRp activity of purified recombinant NS5B proteins, as measured by their abilities to synthesize RNA on an RNA template containing the 3' untranslated region of HCV negative-strand RNA. However, most mutations of the rGTP-specific binding site either impaired or completely ablated the ability of subgenomic HCV RNAs to induce cell colony formation. Likewise, these mutations caused either reduction in or lethality to transient replication of the human immunodeficiency virus Tat-expressing HCV replicon RNAs in the cell. Collectively, these findings demonstrate that the rGTP-specific binding site of the HCV NS5B is not required for in vitro RdRp activity but is important for HCV RNA replication in vivo.
Figures


Similar articles
-
Phosphorylation of hepatitis C virus RNA polymerases ser29 and ser42 by protein kinase C-related kinase 2 regulates viral RNA replication.J Virol. 2014 Oct;88(19):11240-52. doi: 10.1128/JVI.01826-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031343 Free PMC article.
-
Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture.Virology. 2002 Jun 5;297(2):298-306. doi: 10.1006/viro.2002.1461. Virology. 2002. PMID: 12083828
-
Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity.J Virol. 1997 Nov;71(11):8416-28. doi: 10.1128/JVI.71.11.8416-8428.1997. J Virol. 1997. PMID: 9343198 Free PMC article.
-
Functional characterization of fingers subdomain-specific monoclonal antibodies inhibiting the hepatitis C virus RNA-dependent RNA polymerase.J Biol Chem. 2008 Aug 29;283(35):24089-102. doi: 10.1074/jbc.M803422200. Epub 2008 Jun 23. J Biol Chem. 2008. PMID: 18574240 Free PMC article.
-
RNA-dependent RNA polymerase encoded by hepatitis C virus: biomedical applications.Cell Mol Life Sci. 2002 Jun;59(6):909-19. doi: 10.1007/s00018-002-8478-7. Cell Mol Life Sci. 2002. PMID: 12169021 Free PMC article. Review.
Cited by
-
Regulation of de novo-initiated RNA synthesis in hepatitis C virus RNA-dependent RNA polymerase by intermolecular interactions.J Virol. 2010 Jun;84(12):5923-35. doi: 10.1128/JVI.02446-09. Epub 2010 Apr 7. J Virol. 2010. PMID: 20375156 Free PMC article.
-
Structure-function relationships among RNA-dependent RNA polymerases.Curr Top Microbiol Immunol. 2008;320:137-56. doi: 10.1007/978-3-540-75157-1_7. Curr Top Microbiol Immunol. 2008. PMID: 18268843 Free PMC article. Review.
-
Hepatitis C virus NS5B polymerase exhibits distinct nucleotide requirements for initiation and elongation.J Biol Chem. 2008 Dec 5;283(49):33893-901. doi: 10.1074/jbc.M803094200. Epub 2008 Oct 6. J Biol Chem. 2008. PMID: 18840605 Free PMC article.
-
A locking mechanism regulates RNA synthesis and host protein interaction by the hepatitis C virus polymerase.J Biol Chem. 2008 Jul 18;283(29):20535-46. doi: 10.1074/jbc.M801490200. Epub 2008 Apr 28. J Biol Chem. 2008. PMID: 18442978 Free PMC article.
-
Phosphorylation of hepatitis C virus RNA polymerases ser29 and ser42 by protein kinase C-related kinase 2 regulates viral RNA replication.J Virol. 2014 Oct;88(19):11240-52. doi: 10.1128/JVI.01826-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031343 Free PMC article.
References
-
- Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure Fold. Des. 7:1417-1426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

