Sustained CD8+ T-cell responses induced after acute parvovirus B19 infection in humans
- PMID: 16140790
- PMCID: PMC1212640
- DOI: 10.1128/JVI.79.18.12117-12121.2005
Sustained CD8+ T-cell responses induced after acute parvovirus B19 infection in humans
Abstract
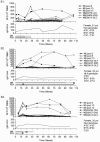
Murine models have suggested that CD8+ T-cell responses peak early in acute viral infections and are not sustained, but no evidence for humans has been available. To address this, we longitudinally analyzed the CD8+ T-cell response to human parvovirus B19 in acutely infected individuals. We observed striking CD8+ T-cell responses, which were sustained or even increased over many months after the resolution of acute disease, indicating that CD8+ T cells may play a prominent role in the control of parvovirus B19 and other acute viral infections of humans, including potentially those generated by live vaccines.
Figures

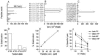
References
-
- Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. - PMC - PubMed
-
- Appay, V., P. R. Dunbar, M. Callan, P. Klenerman, G. M. Gillespie, L. Papagno, G. S. Ogg, A. King, F. Lechner, C. A. Spina, S. Little, D. V. Havlir, D. D. Richman, N. Gruener, G. Pape, A. Waters, P. Easterbrook, M. Salio, V. Cerundolo, A. J. McMichael, and S. L. Rowland-Jones. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8:379-385. - PubMed
-
- Badovinac, V. P., B. B. Porter, and J. T. Harty. 2002. Programmed contraction of CD8(+) T cells after infection. Nat. Immunol. 3:619-626. - PubMed
-
- Franssila, R., and K. Hedman. 2004. T-helper cell-mediated interferon-gamma, interleukin-10 and proliferation responses to a candidate recombinant vaccine for human parvovirus B19. Vaccine 22:3809-3815. - PubMed
-
- Franssila, R., K. Hokynar, and K. Hedman. 2001. T helper cell-mediated in vitro responses of recently and remotely infected subjects to a candidate recombinant vaccine for human parvovirus b19. J. Infect. Dis. 183:805-809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

