Evolutionary mechanisms shaping the genomic structure of the Williams-Beuren syndrome chromosomal region at human 7q11.23
- PMID: 16140988
- PMCID: PMC1199532
- DOI: 10.1101/gr.3944605
Evolutionary mechanisms shaping the genomic structure of the Williams-Beuren syndrome chromosomal region at human 7q11.23
Abstract
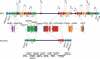
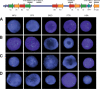
About 5% of the human genome consists of segmental duplications or low-copy repeats, which are large, highly homologous (>95%) fragments of sequence. It has been estimated that these segmental duplications emerged during the past approximately 35 million years (Myr) of human evolution and that they correlate with chromosomal rearrangements. Williams-Beuren syndrome (WBS) is a segmental aneusomy syndrome that is the result of a frequent de novo deletion at 7q11.23, mediated by large (approximately 400-kb) region-specific complex segmental duplications composed of different blocks. We have precisely defined the structure of the segmental duplications on human 7q11.23 and characterized the copy number and structure of the orthologous regions in other primates (macaque, orangutan, gorilla, and chimpanzee). Our data indicate a recent origin and rapid evolution of the 7q11.23 segmental duplications, starting before the diversification of hominoids (approximately 12-16 million years ago [Mya]), with species-specific duplications and intrachromosomal rearrangements that lead to significant differences among those genomes. Alu sequences are located at most edges of the large hominoid-specific segmental duplications, suggesting that they might have facilitated evolutionary rearrangements. We propose a mechanistic model based on Alu-mediated duplicated transposition along with nonallelic homologous recombination for the generation and local expansion of the segmental duplications. The extraordinary rate of evolutionary turnover of this region, rich in segmental duplications, results in important genomic variation among hominoid species, which could be of functional relevance and predispose to disease.
Figures




References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. - PubMed
-
- Armengol, L., Pujana, M.A., Cheung, J., Scherer, S.W., and Estivill, X. 2003. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum. Mol. Genet. 12: 2201-2208. - PubMed
-
- Batzer, M.A. and Deininger, P.L. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370-379. - PubMed
WEB SITE REFERENCES
-
- http://www.ncbi.nlm.nih.gov/; NCBI home page.
-
- http://www.nisc.nih.gov/; The NIH Intramural Sequencing Center (NISC). - PubMed
-
- http://www.ensembl.org/; Ensembl Genome Browser.
-
- http://www.genome.ucsc.edu/; UCSC Genome Browser Home.
-
- http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi; Primer3 Input.
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous
