Transcriptional regulation of phagocytosis-induced membrane biogenesis by sterol regulatory element binding proteins
- PMID: 16141315
- PMCID: PMC1201629
- DOI: 10.1073/pnas.0506716102
Transcriptional regulation of phagocytosis-induced membrane biogenesis by sterol regulatory element binding proteins
Abstract
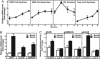
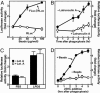
In the process of membrane biogenesis several dozen proteins must operate in precise concert to generate approximately 100 lipids at appropriate concentrations. To study the regulation of bilayer assembly in a cell cycle-independent manner, we have exploited the fact that phagocytes replenish membranes expended during particle engulfment in a rapid phase of lipid synthesis. In response to phagocytosis of latex beads, human embryonic kidney 293 cells synthesized cholesterol and phospholipids at amounts equivalent to the surface area of the internalized particles. Lipid synthesis was accompanied by increased transcription of several lipogenic proteins, including the low-density lipoprotein receptor, enzymes required for cholesterol synthesis (3-hydroxy-3-methylglutaryl CoA synthase, 3-hydroxy-3-methylglutaryl CoA reductase), and fatty acid synthase. Phagocytosis triggered the proteolytic activation of two lipogenic transcription factors, sterol regulatory element binding protein-1a (SREBP-1a) and SREBP-2. Proteolysis of SREBPs coincided with the appearance of their transcriptionally active N termini in the nucleus and 3-fold activation of an SREBP-specific reporter gene. In previous studies with cultured cells, proteolytic activation of SREBP-1a and SREBP-2 has been observed in response to selective starvation of cells for cholesterol and unsaturated fatty acids. However, under the current conditions, SREBP-1a and SREBP-2 are induced without lipid deprivation. SREBP activation is inhibited by high levels of the SREBP-interacting proteins Insig1 or the cytosolic domain of SREBP cleavage-activating protein. Upon overexpression of these proteins, phagocytosis-induced transcription and lipid synthesis were blocked. These results identify SREBPs as essential regulators of membrane biogenesis and provide a useful system for further studies on membrane homeostasis.
Figures




References
-
- Voelker, D. R. (2002) in Biochemistry of Lipids, Lipoproteins and Membranes, eds. Vance, D. E. & Vance, J. E. (Elsevier, Amsterdam), 4th Ed., pp. 449–482.
-
- Tartakoff, A. M., Gordon, S. & Dalbey, R. E., eds. (1999) Phagocytosis: The Host (Elsevier, Amsterdam).
-
- Aderem, A. (2002) Cell 110, 5–8. - PubMed
-
- Werb, Z. & Cohn, Z. A. (1972) J. Biol. Chem. 247, 2439–2446. - PubMed
-
- Sato, R., Yang, J., Wang, X., Evans, M. J., Ho, Y. K., Goldstein, J. L. & Brown, M. S. (1994) J. Biol. Chem. 269, 17267–17273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

