A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells
- PMID: 16141338
- PMCID: PMC1196357
- DOI: 10.1073/pnas.0506306102
A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells
Abstract
The advent of RNA interference has led to the ability to interfere with gene expression and greatly expanded our ability to perform genetic screens in mammalian cells. The expression of short hairpin RNA (shRNA) from polymerase III promoters can be encoded in transgenes and used to produce small interfering RNAs that down-regulate specific genes. In this study, we show that polymerase II-transcribed shRNAs display very efficient knockdown of gene expression when the shRNA is embedded in a microRNA context. Importantly, our shRNA expression system [called PRIME (potent RNA interference using microRNA expression) vectors] allows for the multicistronic cotranscription of a reporter gene, thereby facilitating the tracking of shRNA production in individual cells. Based on this system, we developed a series of lentiviral vectors that display tetracycline-responsive knockdown of gene expression at single copy. The high penetrance of these vectors will facilitate genomewide loss-of-function screens and is an important step toward using bar-coding strategies to follow loss of specific sequences in complex populations.
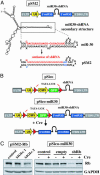
Figures



References
-
- Hannon, G. J. & Rossi, J. J. (2004) Nature 431, 371-378. - PubMed
-
- Silva, J., Chang, K., Hannon, G. J. & Rivas, F. V. (2004) Oncogene 23, 8401-8409. - PubMed
-
- Dykxhoorn, D. M., Novina, C. D. & Sharp, P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 457-467. - PubMed
-
- Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. - PubMed
-
- Mittal, V. (2004) Nat. Rev. Genet. 5, 355-365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

