Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1
- PMID: 16142231
- PMCID: PMC2140048
- DOI: 10.1038/ng1635
Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1
Abstract
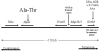
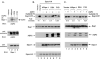
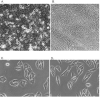
We previously identified loci in the mouse genome that substantially influence the metastatic efficiency of mammary tumors. Here, we present data supporting the idea that the signal transduction molecule, Sipa1, is a candidate for underlying the metastasis efficiency modifier locus Mtes1. Analysis of candidate genes identified a nonsynonymous amino acid polymorphism in Sipa1 that affects the Sipa1 Rap-GAP function. Spontaneous metastasis assays using cells ectopically expressing Sipa1 or cells with knocked-down Sipa1 expression showed that metastatic capacity was correlated with cellular Sipa1 levels. We examined human expression data and found that they were consistent with the idea that Sipa1 concentration has a role in metastasis. Taken together, these data suggest that the Sipa1 polymorphism is one of the genetic polymorphisms underlying the Mtes1 locus. This report is also the first demonstration, to our knowledge, of a constitutional genetic polymorphism affecting tumor metastasis.
Figures







Comment in
-
Metastatic potential as a heritable trait.Nat Genet. 2005 Oct;37(10):1026-7. doi: 10.1038/ng1005-1026. Nat Genet. 2005. PMID: 16195717 No abstract available.
References
-
- Liotta LA, Stetler-Stevenson WG. Principles of molecular cell biology of cancer: Cancer metastasis. J.B. Lippincott Co.; Philadelphia, PA: 1993. pp. 134–149.
-
- Fisher B. From Halsted to prevention and beyond: advances in the management of breast cancer during the twentieth century. Eur J Cancer. 1999;35:1963–73. - PubMed
-
- Heimann R, Lan F, McBride R, Hellman S. Separating favorable from unfavorable prognostic markers in breast cancer: the role of E-cadherin. Cancer Res. 2000;60:298–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
