Catalysis of protein disulfide bond isomerization in a homogeneous substrate
- PMID: 16142915
- PMCID: PMC2526094
- DOI: 10.1021/bi0507985
Catalysis of protein disulfide bond isomerization in a homogeneous substrate
Abstract
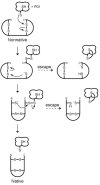
Protein disulfide isomerase (PDI) catalyzes the rearrangement of nonnative disulfide bonds in the endoplasmic reticulum of eukaryotic cells, a process that often limits the rate at which polypeptide chains fold into a native protein conformation. The mechanism of the reaction catalyzed by PDI is unclear. In assays involving protein substrates, the reaction appears to involve the complete reduction of some or all of its nonnative disulfide bonds followed by oxidation of the resulting dithiols. The substrates in these assays are, however, heterogeneous, which complicates mechanistic analyses. Here, we report the first analysis of disulfide bond isomerization in a homogeneous substrate. Our substrate is based on tachyplesin I, a 17-mer peptide that folds into a beta hairpin stabilized by two disulfide bonds. We describe the chemical synthesis of a variant of tachyplesin I in which its two disulfide bonds are in a nonnative state and side chains near its N and C terminus contain a fluorescence donor (tryptophan) and acceptor (N(epsilon)-dansyllysine). Fluorescence resonance energy transfer from 280 to 465 nm increases by 28-fold upon isomerization of the disulfide bonds into their native state (which has a lower E(o') = -0.313 V than does PDI). We use this continuous assay to analyze catalysis by wild-type human PDI and a variant in which the C-terminal cysteine residue within each Cys-Gly-His-Cys active site is replaced with alanine. We find that wild-type PDI catalyzes the isomerization of the substrate with kcat/K(M) = 1.7 x 10(5) M(-1) s(-1), which is the largest value yet reported for catalysis of disulfide bond isomerization. The variant, which is a poor catalyst of disulfide bond reduction and dithiol oxidation, retains virtually all of the activity of wild-type PDI in catalysis of disulfide bond isomerization. Thus, the C-terminal cysteine residues play an insignificant role in the isomerization of the disulfide bonds in nonnative tachyplesin I. We conclude that catalysis of disulfide bond isomerization by PDI does not necessarily involve a cycle of substrate reduction/oxidation.
Figures





References
-
- Thornton JM. Disulphide bridges in globular proteins. J. Mol. Biol. 1981;151:261–287. - PubMed
-
- Darby N, Creighton TE. Disulfide bonds in protein folding and stability. Methods Mol. Biol. 1995;40:219–252. - PubMed
-
- Yano H, Kuroda S, Buchanan BB. Disulfide proteome in the analysis of protein structure and function. Proteomics. 2002;2:1090–1096. - PubMed
-
- Welker E, Wedemeyer WJ, Narayan M, Scheraga HA. Coupling of conformational folding and disulfide-bond reactions in oxidative folding of proteins. Biochemistry. 2001;40:9059–9064. - PubMed
-
- Flory PJ, editor. Statistical Mechanics of Chain Molecules. Wiley; New York, NY: 1969.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

