Molecular recognition via triplex formation of mixed purine/pyrimidine DNA sequences using oligoTRIPs
- PMID: 16144414
- PMCID: PMC2533713
- DOI: 10.1021/ja0530218
Molecular recognition via triplex formation of mixed purine/pyrimidine DNA sequences using oligoTRIPs
Abstract
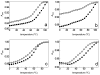
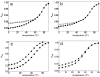
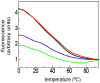
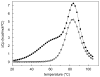
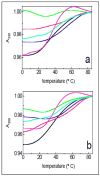
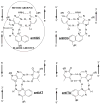
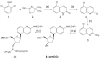
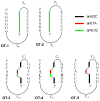
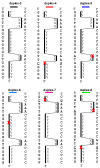
Stable DNA triple-helical structures are normally restricted to homopurine sequences. We have described a system of four heterocyclic bases (TRIPsides) that, when incorporated into oligomers (oligoTRIPs), can recognize and bind in the major groove to any native sequence of DNA [Li et al., J. Am. Chem. Soc. 2003, 125, 2084]. To date, we have reported on triplex-forming oligomers composed of two of these TRIPsides, i.e., antiTA and antiGC, and their ability to form intramolecular triplexes at mixed purine/pyrimidine sequences. In the present study, we describe the synthesis and characterization of the antiCG TRIPside and its use in conjunction with antiTA and antiGC to form sequence-specific intra- and/or intermolecular triplex structures at mixed purine/pyrimidine sequences that require as many as four major groove crossovers.
Figures


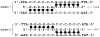
References
-
- Maher LJ, III, Wold B, Dervan PB. Antisense Res Dev. 1991;1:277–281. - PubMed
-
- Praseuth D, Guieysse AL, Hélène C. Biochim Biophys Acta. 1999;1489:181–206. - PubMed
-
- Fox KR. Curr Med Chem. 2000;7:17–37. - PubMed
-
- Dervan PB. Bioorg Med Chem. 2001;9:2215–2235. - PubMed
-
- Kuan JY, Glazer PM. Meth Mol Biol. 2004;262:173–194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

