Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel Staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV
- PMID: 16145133
- PMCID: PMC1234068
- DOI: 10.1128/JCM.43.9.4719-4730.2005
Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel Staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV
Erratum in
- J Clin Microbiol. 2005 Dec;43(12):6223
Abstract
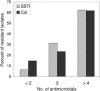
Methicillin-resistant Staphylococcus aureus (MRSA) isolates carry the methicillin resistance gene (mecA) on a horizontally transferred genetic element called the staphylococcal chromosome cassette mec (SCCmec). Community-acquired MRSA (CAMRSA) isolates usually carry SCCmec type IV. We previously reported that 76% of 17 CAMRSA isolates (multilocus sequence type 59) obtained from pediatric patients with skin and soft tissue infections (SSTI) from Taipei did not carry SCCmec types I to IV. We used DNA sequence analysis to determine that the element harbored by these nontypeable isolates is a novel subtype of SCCmec V called SCCmec V(T.) It contains a ccrC recombinase gene variant (ccrC2) and mec complex C2. One SSTI isolate contained molecular features of SCCmec IV but also contained ccrC2 (a feature of SCCmec V(T)), suggesting that it may harbor a composite SCCmec element. The genes lukS-PV and lukF-PV encoding the Panton-Valentine leukocidin (PVL) were present in all CAMRSA SSTI isolates whether they contained SCCmec type IV or V(T). SCCmec V(T) was also present in 5 of 34 (14.7%) CAMRSA colonization isolates collected from healthy children from Taipei who lacked MRSA risk factors. Four (80%) of the these isolates contained lukS-PV and lukF-PV, as did 1 of 27 (3.7%) SCCmec IV-containing colonization isolates. A total of 63% (10 of 16) of the SSTI isolates and 61.7% (21 of 34) of the colonization isolates tested were resistant to at least four classes of non-beta-lactam antimicrobials. SCCmec V(T) is a novel SCCmec variant that is found in multiply resistant CAMRSA strains with sequence type 59 in Taipei in association with the PVL leukotoxin genes.
Figures




References
-
- Adem, P. V., C. P. Montgomery, A. N. Husain, T. K. Koogler, V. Arangelovich, M. Humilier, and R. S. Daum. Novel association of Waterhouse-Friderichsen syndrome with severe Staphylococcus aureus sepsis in children. N. Engl. J. Med., in press. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., Boston, Mass.
-
- Carleton, H. A., B. A. Diep, E. D. Charlebois, G. F. Sensabaugh, and F. Perdreau-Remington. 2004. Community-adapted methicillin-resistant Staphylococcus aureus (MRSA): population dynamics of an expanding community reservoir of MRSA. J. Infect. Dis. 190:1730-1738. - PubMed
-
- CDC. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep. 48:707-710. - PubMed
-
- Chambers, H. F. 2005. Community-associated MRSA: resistance and virulence converge. N. Engl. J. Med. 352:1485-1487. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

