Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression
- PMID: 16148239
- PMCID: PMC6725536
- DOI: 10.1523/JNEUROSCI.1850-05.2005
Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression
Abstract
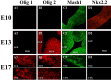
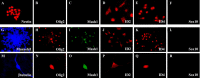
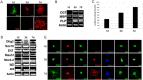
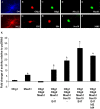
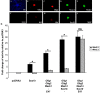
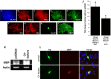
Recent studies suggest that specific neural basic helix-loop-helix (HLH; i.e., Olig1 and Olig2, Mash1), associated inhibitory HLH (i.e., Id2 and Id4), high-mobility group domain (i.e., Sox10), and homeodomain (i.e., Nkx2.2) transcription factors are involved in oligodendrocyte (OL) lineage specification and progressive stages of maturation including myelination. However, the developmental interplay among these lineage-selective determinants, in a cell- and maturational stage-specific context, has not yet been defined. We show here in vivo and in vitro developmental expression profiles for these distinct classes of transcriptional regulators of OLs. We show that progressive stages of OL lineage maturation are characterized by dynamic changes in the subcellular distribution of these transcription factors and by different permutations of combinatorial transcriptional codes. Transient transfections of these precise combinatorial codes with a luciferase reporter gene driven by the myelin basic protein promoter define how changes in the molecular composition of these transcriptional complexes modulate myelin gene expression. Our overall findings suggest that the dynamic interplay between developmental stage-specific classes of transcriptional activators and associated inhibitory factors orchestrate myelin gene expression during terminal maturation of the mammalian CNS.
Figures




Similar articles
-
Differential modulation of BMP signaling promotes the elaboration of cerebral cortical GABAergic neurons or oligodendrocytes from a common sonic hedgehog-responsive ventral forebrain progenitor species.Proc Natl Acad Sci U S A. 2002 Dec 10;99(25):16273-8. doi: 10.1073/pnas.232586699. Epub 2002 Dec 2. Proc Natl Acad Sci U S A. 2002. PMID: 12461181 Free PMC article.
-
Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice.J Neurosci. 2005 Feb 9;25(6):1354-65. doi: 10.1523/JNEUROSCI.3034-04.2005. J Neurosci. 2005. PMID: 15703389 Free PMC article.
-
Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes.J Neurosci. 2007 Dec 26;27(52):14375-82. doi: 10.1523/JNEUROSCI.4456-07.2007. J Neurosci. 2007. PMID: 18160645 Free PMC article.
-
Transcriptional and Epigenetic Regulation of Oligodendrocyte Development and Myelination in the Central Nervous System.Cold Spring Harb Perspect Biol. 2015 Jul 1;7(9):a020461. doi: 10.1101/cshperspect.a020461. Cold Spring Harb Perspect Biol. 2015. PMID: 26134004 Free PMC article. Review.
-
Oligodendrocytes and the control of myelination in vivo: new insights from the rat anterior medullary velum.J Neurosci Res. 2000 Feb 15;59(4):477-88. doi: 10.1002/(SICI)1097-4547(20000215)59:4<477::AID-JNR2>3.0.CO;2-J. J Neurosci Res. 2000. PMID: 10679786 Review.
Cited by
-
Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation.J Neurosci. 2006 Nov 1;26(44):11387-96. doi: 10.1523/JNEUROSCI.3349-06.2006. J Neurosci. 2006. PMID: 17079667 Free PMC article.
-
Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency.Nat Neurosci. 2008 Sep;11(9):1024-34. doi: 10.1038/nn.2172. Nat Neurosci. 2008. PMID: 19160500 Free PMC article.
-
Ascl1/Mash1 promotes brain oligodendrogenesis during myelination and remyelination.J Neurosci. 2013 Jun 5;33(23):9752-9768. doi: 10.1523/JNEUROSCI.0805-13.2013. J Neurosci. 2013. PMID: 23739972 Free PMC article.
-
Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation.PLoS One. 2009 Nov 3;4(11):e7665. doi: 10.1371/journal.pone.0007665. PLoS One. 2009. PMID: 19888342 Free PMC article.
-
Age-related and cuprizone-induced changes in myelin and transcription factor gene expression and in oligodendrocyte cell densities in the rostral corpus callosum of mice.Cell Mol Neurobiol. 2010 May;30(4):607-29. doi: 10.1007/s10571-009-9486-z. Epub 2010 Jan 9. Cell Mol Neurobiol. 2010. PMID: 20063055 Free PMC article.
References
-
- Alberta JA, Song-Kyu P, Mora J, Yuk D-i, Pawlitzky I, Iannarelli P, Vartanian T, Stiles CD, Rowitch DH (2001) Sonic hedgehog is required during an early phase of oligodendrocyte development in mammalian brain. Mol Cell Neurosci 18: 434–441. - PubMed
-
- Durand B, Raff M (2000) A cell-intrinsic timer that operates during oligodendrocyte development. BioEssays 22: 64–71. - PubMed
-
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J (1997) Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90: 169–180. - PubMed
-
- Gunhaga L, Jessell TM, Edlund T (2000) Sonic hedgehog signaling at gastrula stages specifies ventral telencephalic cell in the chick embryo. Development 127: 3283–3293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
