N-ethyl-N-nitrosourea mutagenesis: boarding the mouse mutant express
- PMID: 16148305
- PMCID: PMC1197804
- DOI: 10.1128/MMBR.69.3.426-439.2005
N-ethyl-N-nitrosourea mutagenesis: boarding the mouse mutant express
Abstract
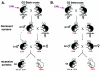
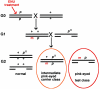
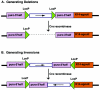
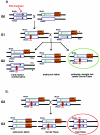
In the mouse, random mutagenesis with N-ethyl-N-nitrosourea (ENU) has been used since the 1970s in forward mutagenesis screens. However, only in the last decade has ENU mutagenesis been harnessed to generate a myriad of new mouse mutations in large-scale genetic screens and focused, smaller efforts. The development of additional genetic tools, such as balancer chromosomes, refinements in genetic mapping strategies, and evolution of specialized assays, has allowed these screens to achieve new levels of sophistication. The impressive productivity of these screens has led to a deluge of mouse mutants that wait to be harnessed. Here the basic large- and small-scale strategies are described, as are the basics of screen design. Finally, and importantly, this review describes the mechanisms by which such mutants may be accessed now and in the future. Thus, this review should serve both as an overview of the power of forward mutagenesis in the mouse and as a resource for those interested in developing their own screens, adding onto existing efforts, or obtaining specific mouse mutants that have already been generated.
Figures
References
-
- Abbott, A. 2004. Geneticists prepare for deluge of mutant mice. Nature 432:541. - PubMed
-
- Anderson, K. V. 2000. Finding the genes that direct mammalian development: ENU mutagenesis in the mouse. Trends Genet. 16:99-102. - PubMed
-
- Brown, S. D., and R. Balling. 2001. Systematic approaches to mouse mutagenesis. Curr. Opin. Genet. Dev. 11:268-273. - PubMed
-
- Carpinelli, M. R., D. J. Hilton, D. Metcalf, J. L. Antonchuk, C. D. Hyland, S. L. Mifsud, L. Di Rago, A. A. Hilton, T. A. Willson, A. W. Roberts, R. G. Ramsay, N. A. Nicola, and W. Alexander, S. 2004. Suppressor screen in Mpl−/− mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc. Natl. Acad. Sci. USA 101:6553-6558. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

