Replication termination in Escherichia coli: structure and antihelicase activity of the Tus-Ter complex
- PMID: 16148308
- PMCID: PMC1197808
- DOI: 10.1128/MMBR.69.3.501-526.2005
Replication termination in Escherichia coli: structure and antihelicase activity of the Tus-Ter complex
Abstract
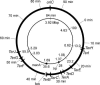
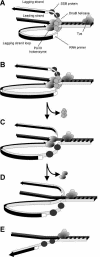
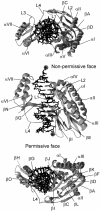
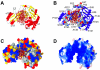
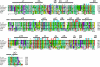
The arrest of DNA replication in Escherichia coli is triggered by the encounter of a replisome with a Tus protein-Ter DNA complex. A replication fork can pass through a Tus-Ter complex when traveling in one direction but not the other, and the chromosomal Ter sites are oriented so replication forks can enter, but not exit, the terminus region. The Tus-Ter complex acts by blocking the action of the replicative DnaB helicase, but details of the mechanism are uncertain. One proposed mechanism involves a specific interaction between Tus-Ter and the helicase that prevents further DNA unwinding, while another is that the Tus-Ter complex itself is sufficient to block the helicase in a polar manner, without the need for specific protein-protein interactions. This review integrates three decades of experimental information on the action of the Tus-Ter complex with information available from the Tus-TerA crystal structure. We conclude that while it is possible to explain polar fork arrest by a mechanism involving only the Tus-Ter interaction, there are also strong indications of a role for specific Tus-DnaB interactions. The evidence suggests, therefore, that the termination system is more subtle and complex than may have been assumed. We describe some further experiments and insights that may assist in unraveling the details of this fascinating process.
Figures
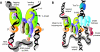


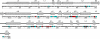


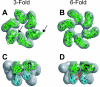
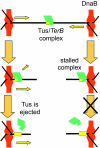

References
-
- Abhyankar, M. M., J. M. Reddy, R. Sharma, E. Büllesbach, and D. Bastia. 2004. Biochemical investigations of the control of replication initiation of plasmid R6K. J. Biol. Chem. 279:6711-6719. - PubMed
-
- Abhyankar, M. M., S. Zzaman, and D. Bastia. 2003. Reconstitution of R6K DNA replication in vitro using 22 purified proteins. J. Biol. Chem. 278:45476-45484. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Amin, A. A., and J. Hurwitz. 1992. Polar arrest of the simian virus 40 tumor antigen-mediated replication fork movement in vitro by the tus protein-terB complex of Escherichia coli. J. Biol. Chem. 267:18612-18622. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

