Colonization of cattle intestines by Campylobacter jejuni and Campylobacter lanienae
- PMID: 16151098
- PMCID: PMC1214653
- DOI: 10.1128/AEM.71.9.5145-5153.2005
Colonization of cattle intestines by Campylobacter jejuni and Campylobacter lanienae
Abstract
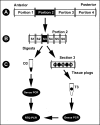
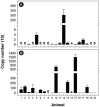
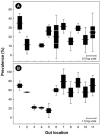
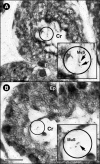
The location and abundance of Campylobacter jejuni and Campylobacter lanienae in the intestines of beef cattle were investigated using real-time quantitative PCR in two studies. In an initial study, digesta and tissue samples were obtained along the digestive tract of two beef steers known to shed C. jejuni and C. lanienae (steers A and B). At the time of slaughter, steer B weighed 540 kg, compared to 600 kg for steer A, yet the intestine of steer B (40.5 m) was 36% longer than the intestine of steer A (26.1 m). In total, 323 digesta samples (20-cm intervals) and 998 tissue samples (3.3- to 6.7-cm intervals) were processed. Campylobacter DNA was detected in the digesta and in association with tissues throughout the small and large intestines of both animals. Although C. jejuni and C. lanienae DNA were detected in both animals, only steer A contained substantial quantities of C. jejuni DNA. In both digesta and tissues of steer A, C. jejuni was present in the duodenum and jejunum. Considerable quantities of C. jejuni DNA also were observed in the digesta obtained from the cecum and ascending colon, but minimal DNA was associated with tissues of these regions. In contrast, steer B contained substantial quantities of C. lanienae DNA, and DNA of this bacterium was limited to the large intestine (i.e., the cecum, proximal ascending colon, descending colon, and rectum); the majority of tissue-associated C. lanienae DNA was present in the cecum, descending colon, and rectum. In a second study, the location and abundance of C. jejuni and C. lanienae DNA were confirmed in the intestines of 20 arbitrarily selected beef cattle. DNA of C. jejuni and C. lanienae were detected in the digesta of 57% and 95% of the animals, respectively. C. jejuni associated with intestinal tissues was most abundant in the duodenum, ileum, and rectum. However, one animal contributed disproportionately to the abundance of C. jejuni DNA in the ileum and rectum. C. lanienae was most abundant in the large intestine, and the highest density of DNA of this bacterium was found in the cecum. Therefore, C. jejuni colonized the proximal small intestine of asymptomatic beef cattle, whereas C. lanienae primarily resided in the cecum, descending colon, and rectum. This information could be instrumental in developing efficacious strategies to manage the release of these bacteria from the gastrointestinal tracts of cattle.
Figures


References
-
- Al-Mashat, R. R., and D. J. Taylor. 1980. Campylobacter spp. in enteric lesions in cattle. Vet. Rec. 107:31-34. - PubMed
-
- Al-Mashat, R. R., and D. J. Taylor. 1980. Production of diarrhoea and dysentery in experimental calves by feeding pure cultures of Campylobacter fetus subspecies jejuni. Vet. Rec. 107:459-464. - PubMed
-
- Al-Mashat, R. R., and D. J. Taylor. 1981. Production of enteritis in calves by the oral inoculation of pure cultures of Campylobacter fecalis. Vet. Rec. 109:97-101. - PubMed
-
- Al-Mashat, R. R., and D. J. Taylor. 1983. Bacteria in enteric lesions of cattle. Vet. Rec. 112:5-10. - PubMed
-
- Al-Mashat, R. R., and D. J. Taylor. 1983. Production of enteritis in calves by the oral inoculation of pure cultures of Campylobacter fetus subspecies intestinalis. Vet. Rec. 112:54-58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

