Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean
- PMID: 16151126
- PMCID: PMC1214674
- DOI: 10.1128/AEM.71.9.5362-5370.2005
Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean
Abstract
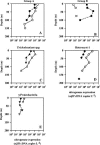
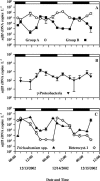
Dinitrogen (N(2))-fixing microorganisms (diazotrophs) play important roles in ocean biogeochemistry and plankton productivity. In this study, we examined the presence and expression of specific planktonic nitrogenase genes (nifH) in the upper ocean (0 to 175 m) at Station ALOHA in the oligotrophic North Pacific Ocean. Clone libraries constructed from reverse-transcribed PCR-amplified mRNA revealed six unique phylotypes. Five of the nifH phylotypes grouped with sequences from unicellular and filamentous cyanobacteria, and one of the phylotypes clustered with gamma-proteobacteria. The cyanobacterial nifH phylotypes retrieved included two sequence types that phylogenetically grouped with unicellular cyanobacteria (termed groups A and B), several sequences closely related (97 to 99%) to Trichodesmium spp. and Katagnymene spiralis, and two previously unreported phylotypes clustering with heterocyst-forming nifH cyanobacteria. Temporal patterns of nifH expression were evaluated using reverse-transcribed quantitative PCR amplification of nifH gene transcripts. The filamentous and presumed unicellular group A cyanobacterial phylotypes exhibited elevated nifH transcription during the day, while members of the group B (closely related to Crocosphaera watsonii) unicellular phylotype displayed greater nifH transcription at night. In situ nifH expression by all of the cyanobacterial phylotypes exhibited pronounced diel periodicity. The gamma-proteobacterial phylotype had low transcript abundance and did not exhibit a clear diurnal periodicity in nifH expression. The temporal separation of nifH expression by the various phylotypes suggests that open ocean diazotrophic cyanobacteria have unique in situ physiological responses to daily fluctuations of light in the upper ocean.
Figures


References
-
- Braun, S. T., L. M. Proctor, S. Zani, M. T. Mellon, and J. P. Zehr. 1999. Molecular evidence for zooplankton-associated nitrogen-fixing anaerobes based on amplification of the nifH gene. FEMS Microbiol. Lett. 28:273-279.
-
- Capone, D. G., and E. J. Carpenter. 1982. Nitrogen fixation in the marine environment. Science 217:1140-1142. - PubMed
-
- Capone, D. G., J. P. Zehr, H. W. Paerl, B. Bergman, and E. J. Carpenter. 1997. Trichodesmium, a globally significant marine cyanobacterium. Science 276:1221-1229.
-
- Carpenter, E. J. 1983. Nitrogen fixation by marine Oscillatoria (Trichodesmium) in the world's oceans, p. 65-103. In E. J. Carpenter and D. G. Capone (ed.), Nitrogen in the marine environment. Academic Press, Inc., New York, N.Y.
-
- Carpenter, E. J., and S. Janson. 2001. Anabaena gerdii sp. nov., a new planktonic filamentous cyanobacterium from the South Pacific Ocean and Arabian Sea. Phycologia 40:105-110.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

