Design and evaluation of 16S rRNA-targeted peptide nucleic acid probes for whole-cell detection of members of the genus Listeria
- PMID: 16151137
- PMCID: PMC1214657
- DOI: 10.1128/AEM.71.9.5451-5457.2005
Design and evaluation of 16S rRNA-targeted peptide nucleic acid probes for whole-cell detection of members of the genus Listeria
Abstract
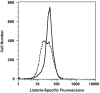
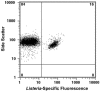
Six fluorescein-labeled peptide nucleic acid oligomers targeting Listeria-specific sequences on the 16S ribosomal subunit were evaluated for their abilities to hybridize to whole cells by fluorescence in situ hybridization (FISH). Four of these probes yielded weak or no fluorescent signals after hybridization and were not investigated further. The remaining two FISH-compatible probes, LisUn-3 and LisUn-11, were evaluated for their reactivities against 22 Listeria strains and 17 other bacterial strains belonging to 10 closely related genera. Hybridization with BacUni-1, a domain-specific eubacterial probe, was used as a positive control for target accessibility in both Listeria spp. and nontarget cells. RNase T1 treatment of select cell types was used to confirm that positive fluorescence responses were rRNA dependent and to examine the extent of nonspecific staining of nontarget cells. Both LisUn-3 and LisUn-11 yielded rapid, bright, and genus-specific hybridizations at probe concentrations of approximately 100 pmol ml(-1). LisUn-11 was the brightest probe and stained all six Listeria species. LisUn-3 hybridized with all Listeria spp. except for L. grayi, for which it had two mismatched bases. A simple ethanolic fixation yielded superior results with Listeria spp. compared to fixation in 10% buffered formalin and was applicable to all cell types studied. This study highlights the advantages of peptide nucleic acid probes for FISH-based detection of gram-positive bacteria and provides new tools for the rapid detection of Listeria spp. These probes may be useful for the routine monitoring of food production environments in support of efforts to control L. monocytogenes.
Figures


References
-
- Anderson, M. L. M. 1999. Solution hybridization: reaction conditions, p. 51. In Nucleic acid hybridization. BIOS Scientific Publishers, Oxford, United Kingdom.
-
- Bohnert, J., B. Hübner, and K. Botzenhart. 2000. Rapid identification of Enterobacteriaceae using a novel 23S rRNA-targeted oligonucleotide probe. Int. J. Hyg. Environ. Health 203:77-82. - PubMed
-
- Collins, M. D., S. Wallbanks, D. J. Lane, J. Shah, R. Nietupski, J. Smida, M. Dorsch, and E. Stackebrandt. 1991. Phylogenetic analysis of the genus Listeria based on reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:240-246. - PubMed
-
- Federal Register. 2001. Performance standards for the production of processed meat and poultry products. Fed. Regist. 66:12590-12636.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

