Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states
- PMID: 16154092
- PMCID: PMC3685586
- DOI: 10.1016/j.str.2005.06.006
Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states
Abstract
Response regulators (RRs), which undergo phosphorylation/dephosphorylation at aspartate residues, are highly prevalent in bacterial signal transduction. RRs typically contain an N-terminal receiver domain that regulates the activities of a C-terminal DNA binding domain in a phosphorylation-dependent manner. We present crystallography and solution NMR data for the receiver domain of Escherichia coli PhoB which show distinct 2-fold symmetric dimers in the inactive and active states. These structures, together with the previously determined structure of the C-terminal domain of PhoB bound to DNA, define the conformation of the active transcription factor and provide a model for the mechanism of activation in the OmpR/PhoB subfamily, the largest group of RRs. In the active state, the receiver domains dimerize with 2-fold rotational symmetry using their alpha4-beta5-alpha5 faces, while the effector domains bind to DNA direct repeats with tandem symmetry, implying a loss of intramolecular interactions.
Figures

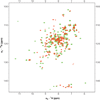



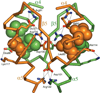
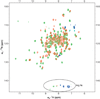
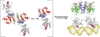
Comment in
-
No phobias about PhoB activation.Structure. 2005 Sep;13(9):1238-9. doi: 10.1016/j.str.2005.08.002. Structure. 2005. PMID: 16154079
References
-
- Aiba H, Mizuno T. Phosphorylation of a bacterial activator protein, OmpR, by a protein kinase, EnvZ, stimulates the transcription of the ompF and ompC genes in Escherichia coli. FEBS Lett. 1990;261:19–22. - PubMed
-
- Aoyama T, Takanami M, Makino K, Oka A. Cross-talk between the virulence and phosphate regulons of Agrobacterium tumefaciens caused by an unusual interaction of the transcriptional activator with a regulatory DNA element. Mol. Gen. Genet. 1991;227:385–390. - PubMed
-
- Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus RP, Dickerson RE. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. - PubMed
-
- Bellsolell L, Prieto J, Serrano L, Coll M. Magnesium binding to the bacterial chemotaxis protein CheY results in large conformational changes involving its functional surface. J. Mol. Biol. 1994;238:489–495. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

