SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis
- PMID: 16155178
- PMCID: PMC1242263
- DOI: 10.1105/tpc.105.035485
SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis
Abstract
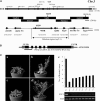
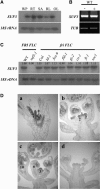
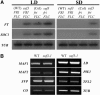
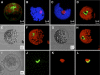
Flowering traits in winter annual Arabidopsis thaliana are conferred mainly by two genes, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC). FLC acts as a flowering repressor and is regulated by multiple flowering pathways. We isolated an early-flowering mutant, suppressor of FRIGIDA3 (suf3), which also shows leaf serration, weak apical dominance, and infrequent conversion of the inflorescence shoot to a terminal flower. The suf3 mutation caused a decrease in the transcript level of FLC in both a FRI-containing line and autonomous pathway mutants. However, suf3 showed only a partial reduction of FLC transcript level, although it largely suppressed the late-flowering phenotype. In addition, the suf3 mutation caused acceleration of flowering in both 35S-FLC and a flc null mutant, indicating that SUF3 regulates additional factor(s) for the repression of flowering. SUF3 is highly expressed in the shoot apex, but the expression is not regulated by FRI, autonomous pathway genes, or vernalization. SUF3 encodes the nuclear ACTIN-RELATED PROTEIN6 (ARP6), the homolog of which in yeast is a component of an ATP-dependent chromatin-remodeling SWR1 complex. Our analyses showed that SUF3 regulates FLC expression independent of vernalization, FRI, and an autonomous pathway gene, all of which affect the histone modification of FLC chromatin. Subcellular localization using a green fluorescent protein fusion showed that Arabidopsis ARP6 is located at distinct regions of the nuclear periphery.
Figures



References
-
- Amasino, R. (2004). Take a cold flower. Nat. Genet. 36, 111–112. - PubMed
-
- Ausin, I., Alonso-Blanco, C., Jarillo, J.A., Ruiz-Garcia, L., and Martinez-Zapater, J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36, 162–166. - PubMed
-
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. - PubMed
-
- Blessing, C.A., Ugrinova, G.T., and Goodson, H.V. (2004). Actin and ARPs: Action in the nucleus. Trends Cell Biol. 14, 435–442. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

