Constitutive expression of catABC genes in the aniline-assimilating bacterium Rhodococcus species AN-22: production, purification, characterization and gene analysis of CatA, CatB and CatC
- PMID: 16156722
- PMCID: PMC1383680
- DOI: 10.1042/BJ20050740
Constitutive expression of catABC genes in the aniline-assimilating bacterium Rhodococcus species AN-22: production, purification, characterization and gene analysis of CatA, CatB and CatC
Abstract
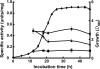
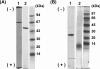
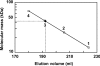
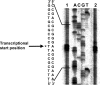
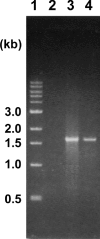
The aniline-assimilating bacterium Rhodococcus sp. AN-22 was found to constitutively synthesize CatB (cis,cis-muconate cycloisomerase) and CatC (muconolactone isomerase) in its cells growing on non-aromatic substrates, in addition to the previously reported CatA (catechol 1,2-dioxygenase). The bacterium maintained the specific activity of the three enzymes at an almost equal level during cultivation on succinate. CatB and CatC were purified to homogeneity and characterized. CatB was a monomer with a molecular mass of 44 kDa. The enzyme was activated by Mn2+, Co2+ and Mg2+. Native CatC was a homo-octamer with a molecular mass of 100 kDa. The enzyme was stable between pH 7.0 and 10.5 and was resistant to heating up to 90 degrees C. Genes coding for CatA, CatB and CatC were cloned and named catA, catB and catC respectively. The catABC genes were transcribed as one operon. The deduced amino acid sequences of CatA, CatB and CatC showed high identities with those from other Gram-positive micro-organisms. A regulator gene such as catR encoding a regulatory protein was not observed around the cat gene cluster of Rhodococcus sp. AN-22, but a possible relic of catR was found in the upstream region of catA. Reverse transcriptase-PCR and primer extension analyses showed that the transcriptional start site of the cat gene cluster was located 891 bp upstream of the catA initiation codon in the AN-22 strain growing on both aniline and succinate. Based on these data, we concluded that the bacterium constitutively transcribed the catABC genes and translated its mRNA into CatA, CatB and CatC.
Figures
References
-
- Aoki K., Shinke R., Nishira H. Identification of aniline-assimilating bacteria. Agric. Biol. Chem. 1982;46:2563–2570.
-
- Aoki K., Shinke R., Nishira H. Metabolism of aniline by Rhodococcus erythropolis AN-13. Agric. Biol. Chem. 1983;48:1611–1616.
-
- Aoki K., Konohana T., Shinke R., Nishira H. Purification and characterization of catechol 1,2-dioxygenase from aniline-assimilating Rhodococcus erythropolis AN-13. Agric. Biol. Chem. 1984;48:2087–2095.
-
- Aoki K., Konohana T., Shinke R., Nishira H. Two catechol 1,2-dioxygenases from an aniline-assimilating bacterium, Frateuria sp. ANA-18. Agric. Biol. Chem. 1984;48:2097–2104.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

