Duplication and positive selection among hominin-specific PRAME genes
- PMID: 16159394
- PMCID: PMC1262708
- DOI: 10.1186/1471-2164-6-120
Duplication and positive selection among hominin-specific PRAME genes
Abstract
Background: The physiological and phenotypic differences between human and chimpanzee are largely specified by our genomic differences. We have been particularly interested in recent duplications in the human genome as examples of relatively large-scale changes to our genome. We performed an in-depth evolutionary analysis of a region of chromosome 1, which is copy number polymorphic among humans, and that contains at least 32 PRAME (Preferentially expressed antigen of melanoma) genes and pseudogenes. PRAME-like genes are expressed in the testis and in a large number of tumours, and are thought to possess roles in spermatogenesis and oogenesis.
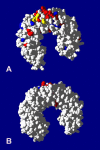
Results: Using nucleotide substitution rate estimates for exons and introns, we show that two large segmental duplications, of six and seven human PRAME genes respectively, occurred in the last 3 million years. These duplicated genes are thus hominin-specific, having arisen in our genome since the divergence from chimpanzee. This cluster of PRAME genes appears to have arisen initially from a translocation approximately 95-85 million years ago. We identified multiple sites within human or mouse PRAME sequences which exhibit strong evidence of positive selection. These form a pronounced cluster on one face of the predicted PRAME protein structure.
Conclusion: We predict that PRAME genes evolved adaptively due to strong competition between rapidly-dividing cells during spermatogenesis and oogenesis. We suggest that as PRAME gene copy number is polymorphic among individuals, positive selection of PRAME alleles may still prevail within the human population.
Figures





References
-
- Brunet M, Guy F, Pilbeam D, Mackaye HT, Likius A, Ahounta D, Beauvilain A, Blondel C, Bocherens H, Boisserie JR, De Bonis L, Coppens Y, Dejax J, Denys C, Duringer P, Eisenmann V, Fanone G, Fronty P, Geraads D, Lehmann T, Lihoreau F, Louchart A, Mahamat A, Merceron G, Mouchelin G, Otero O, Pelaez Campomanes P, Ponce De Leon M, Rage JC, Sapanet M, Schuster M, Sudre J, Tassy P, Valentin X, Vignaud P, Viriot L, Zazzo A, Zollikofer C. A new hominid from the Upper Miocene of Chad, Central Africa. Nature. 2002;418:145–151. doi: 10.1038/nature00879. - DOI - PubMed
-
- Winter H, Langbein L, Krawczak M, Cooper DN, Jave-Suarez LF, Rogers MA, Praetzel S, Heidt PJ, Schweizer J. Human type I hair keratin pseudogene phihHaA has functional orthologs in the chimpanzee and gorilla: evidence for recent inactivation of the human gene after the Pan-Homo divergence. Hum Genet. 2001;108:37–42. doi: 10.1007/s004390000439. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

