Functional genomics and expression analysis of the Corynebacterium glutamicum fpr2-cysIXHDNYZ gene cluster involved in assimilatory sulphate reduction
- PMID: 16159395
- PMCID: PMC1266029
- DOI: 10.1186/1471-2164-6-121
Functional genomics and expression analysis of the Corynebacterium glutamicum fpr2-cysIXHDNYZ gene cluster involved in assimilatory sulphate reduction
Abstract
Background: Corynebacterium glutamicum is a high-GC Gram-positive soil bacterium of great biotechnological importance for the production of amino acids. To facilitate the rational design of sulphur amino acid-producing strains, the pathway for assimilatory sulphate reduction providing the necessary reduced sulfur moieties has to be known. Although this pathway has been well studied in Gram-negative bacteria like Escherichia coli and low-GC Gram-positives like Bacillus subtilis, little is known for the Actinomycetales and other high-GC Gram-positive bacteria.
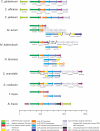
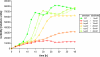
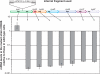
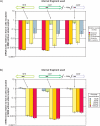
Results: The genome sequence of C. glutamicum was searched for genes involved in the assimilatory reduction of inorganic sulphur compounds. A cluster of eight candidate genes could be identified by combining sequence similarity searches with a subsequent synteny analysis between C. glutamicum and the closely related C. efficiens. Using mutational analysis, seven of the eight candidate genes, namely cysZ, cysY, cysN, cysD, cysH, cysX, and cysI, were demonstrated to be involved in the reduction of inorganic sulphur compounds. For three of the up to now unknown genes possible functions could be proposed: CysZ is likely to be the sulphate permease, while CysX and CysY are possibly involved in electron transfer and cofactor biosynthesis, respectively. Finally, the candidate gene designated fpr2 influences sulphur utilisation only weakly and might be involved in electron transport for the reduction of sulphite. Real-time RT-PCR experiments revealed that cysIXHDNYZ form an operon and that transcription of the extended cluster fpr2 cysIXHDNYZ is strongly influenced by the availability of inorganic sulphur, as well as L-cysteine. Mapping of the fpr2 and cysIXHDNYZ promoters using RACE-PCR indicated that both promoters overlap with binding-sites of the transcriptional repressor McbR, suggesting an involvement of McbR in the observed regulation. Comparative genomics revealed that large parts of the extended cluster are conserved in 11 of 17 completely sequenced members of the Actinomycetales.
Conclusion: The set of C. glutamicum genes involved in assimilatory sulphate reduction was identified and four novel genes involved in this pathway were found. The high degree of conservation of this cluster among the Actinomycetales supports the hypothesis that a different metabolic pathway for the reduction of inorganic sulphur compounds than that known from the well-studied model organisms E. coli and B. subtilis is used by members of this order, providing the basis for further biochemical studies.
Figures


References
-
- Kredich NM. Biosynthesis of Cysteine. In: Neidhardt FC, Curtis III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2. Vol. 2. Washington DC: ASM Press; 1996. pp. 514–527.
-
- Mansilla MC, de Mendoza D. The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (Pit) family. Microbiology. 2000;146:815–821. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

