Positive autoregulation of cI is a dispensable feature of the phage lambda gene regulatory circuitry
- PMID: 16159777
- PMCID: PMC1236637
- DOI: 10.1128/JB.187.18.6430-6442.2005
Positive autoregulation of cI is a dispensable feature of the phage lambda gene regulatory circuitry
Abstract
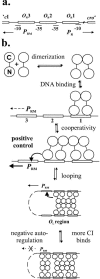
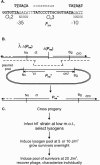
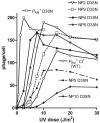
Complex gene regulatory circuits contain many features that are likely to contribute to their operation. It is unclear, however, whether all these features are necessary for proper circuit behavior or whether certain ones are refinements that make the circuit work better but are dispensable for qualitatively normal behavior. We have addressed this question using the phage lambda regulatory circuit, which can persist in two stable states, the lytic state and the lysogenic state. In the lysogenic state, the CI repressor positively regulates its own expression by stimulating transcription from the P(RM) promoter. We tested whether this feature is an essential part of the regulatory circuitry. Several phages with a cI mutation preventing positive autoregulation and an up mutation in the P(RM) promoter showed near-normal behavior. We conclude that positive autoregulation is not necessary for proper operation of the lambda circuitry and speculate that it serves a partially redundant function of stabilizing a bistable circuit, a form of redundancy we term "circuit-level redundancy." We discuss our findings in the context of a two-stage model for evolution and elaboration of regulatory circuits from simpler to more complex forms.
Figures
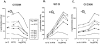

References
-
- Bailone, A., A. Levine, and R. Devoret. 1979. Inactivation of prophage λ repressor in vivo. J. Mol. Biol. 131:553-572. - PubMed
-
- Bushman, F. D., C. Shang, and M. Ptashne. 1989. A single glutamic acid residue plays a key role in the transcriptional activation function of lambda repressor. Cell 58:1163-1171. - PubMed
-
- Darling, P. J., J. M. Holt, and G. K. Ackers. 2000. Coupled energetics of lambda cro repressor self-assembly and site-specific DNA operator binding I: analysis of cro dimerization from nanomolar to micromolar concentrations. Biochemistry 39:11500-11507. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

