Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis
- PMID: 16160147
- PMCID: PMC1211555
- DOI: 10.1128/JVI.79.19.12205-12217.2005
Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis
Abstract
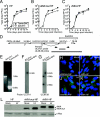
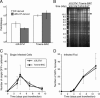
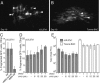
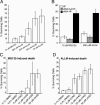
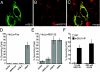
Human cytomegalovirus carries a mitochondria-localized inhibitor of apoptosis (vMIA) that is conserved in primate cytomegaloviruses. We find that inactivating mutations within UL37x1, which encodes vMIA, do not substantially affect replication in TownevarATCC (Towne-BAC), a virus that carries a functional copy of the betaherpesvirus-conserved viral inhibitor of caspase 8 activation, the UL36 gene product. In Towne-BAC infection, vMIA reduces susceptibility of infected cells to intrinsic death induced by proteasome inhibition. vMIA is sufficient to confer resistance to proteasome inhibition when expressed independent of viral infection. Murine cytomegalovirus m38.5, whose position in the viral genome is analogous to UL37x1, exhibits mitochondrial association and functions in much the same manner as vMIA in inhibiting intrinsic cell death. This work suggests a common role for vMIA in rodent and primate cytomegaloviruses, modulating the threshold of virus-infected cells to intrinsic cell death.
Figures

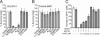
Similar articles
-
Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses.Virology. 2003 Nov 25;316(2):221-33. doi: 10.1016/j.virol.2003.07.003. Virology. 2003. PMID: 14644605
-
vMIA, a viral inhibitor of apoptosis targeting mitochondria.Biochimie. 2002 Feb-Mar;84(2-3):177-85. doi: 10.1016/s0300-9084(02)01367-6. Biochimie. 2002. PMID: 12022948 Review.
-
Cytopathic effects of the cytomegalovirus-encoded apoptosis inhibitory protein vMIA.J Cell Biol. 2006 Sep 25;174(7):985-96. doi: 10.1083/jcb.200604069. Epub 2006 Sep 18. J Cell Biol. 2006. PMID: 16982800 Free PMC article.
-
HtrA2/Omi terminates cytomegalovirus infection and is controlled by the viral mitochondrial inhibitor of apoptosis (vMIA).PLoS Pathog. 2008 May 9;4(5):e1000063. doi: 10.1371/journal.ppat.1000063. PLoS Pathog. 2008. PMID: 18769594 Free PMC article.
-
Cell death suppression by cytomegaloviruses.Apoptosis. 2005 Mar;10(2):251-65. doi: 10.1007/s10495-005-0800-z. Apoptosis. 2005. PMID: 15843887 Review.
Cited by
-
Human Cytomegalovirus pUL37x1 Is Important for Remodeling of Host Lipid Metabolism.J Virol. 2019 Oct 15;93(21):e00843-19. doi: 10.1128/JVI.00843-19. Print 2019 Nov 1. J Virol. 2019. PMID: 31391267 Free PMC article.
-
Regulation of the subcellular distribution of key cellular RNA-processing factors during permissive human cytomegalovirus infection.J Gen Virol. 2010 Jun;91(Pt 6):1547-59. doi: 10.1099/vir.0.020313-0. Epub 2010 Feb 17. J Gen Virol. 2010. PMID: 20164265 Free PMC article.
-
Contribution of GADD45 family members to cell death suppression by cellular Bcl-xL and cytomegalovirus vMIA.J Virol. 2005 Dec;79(23):14923-32. doi: 10.1128/JVI.79.23.14923-14932.2005. J Virol. 2005. PMID: 16282491 Free PMC article.
-
A novel inhibitory mechanism of mitochondrion-dependent apoptosis by a herpesviral protein.PLoS Pathog. 2007 Dec;3(12):e174. doi: 10.1371/journal.ppat.0030174. PLoS Pathog. 2007. PMID: 18069888 Free PMC article.
-
Viral and cell cycle-regulated kinases in cytomegalovirus-induced pseudomitosis and replication.PLoS Pathog. 2007 Jan;3(1):e6. doi: 10.1371/journal.ppat.0030006. PLoS Pathog. 2007. PMID: 17206862 Free PMC article.
References
-
- Arnoult, D., L. Bartle, A. Skaletskaya, D. Poncet, N. Zamzami, P. Park, J. Sharpe, R. Youle, and V. S. Goldmacher. 2004. Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. USA 101:7988-7993. - PMC - PubMed
-
- Bartz, S., and M. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

