Four-dimensional visualization of the simultaneous activity of alternative adeno-associated virus replication origins
- PMID: 16160148
- PMCID: PMC1211535
- DOI: 10.1128/JVI.79.19.12218-12230.2005
Four-dimensional visualization of the simultaneous activity of alternative adeno-associated virus replication origins
Abstract
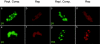
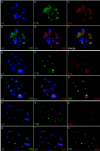
The adeno-associated virus (AAV) inverted terminal repeats (ITRs) contain the AAV Rep protein-binding site (RBS) and the terminal resolution site (TRS), which together act as a minimal origin of DNA replication. The AAV p5 promoter also contains an RBS, which is involved in Rep-mediated regulation of promoter activity, as well as a functional TRS, and origin activity of these signals has in fact been demonstrated previously in the presence of adenovirus helper functions. Here, we show that in the presence of herpes simplex virus type 1 (HSV-1) and AAV Rep protein, p5 promoter-bearing plasmids are efficiently amplified to form large head-to-tail concatemers, which are readily packaged in HSV-1 virions if an HSV-1 DNA-packaging/cleavage signal is provided in cis. We also demonstrate simultaneous and independent replication from the two alternative AAV replication origins, p5 and ITR, on the single-cell level using multicolor-fluorescence live imaging, a finding which raises the possibility that both origins may contribute to the AAV life cycle. Furthermore, we assess the differential affinities of Rep for the two different replication origins, p5 and ITR, both in vitro and in live cells and identify this as a potential mechanism to control the replicative and promoter activities of p5.
Figures





References
-
- Caleo, M., M. C. Cenni, M. Costa, E. Menna, L. Zentilin, S. Giadrossi, M. Giacca, and L. Maffei. 2002. Expression of BCL-2 via adeno-associated virus vectors rescues thalamic neurons after visual cortex lesion in the adult rat. Eur. J. Neurosci. 15:1271-1277. - PubMed
-
- Chadeuf, G., D. Favre, J. Tessier, N. Provost, P. Nony, J. Kleinschmidt, P. Moullier, and A. Salvetti. 2000. Efficient recombinant adeno-associated virus production by a stable rep-cap HeLa cell line correlates with adenovirus-induced amplification of the integrated rep-cap genome. J. Gene Med. 2:260-268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

