MITF and cell proliferation: the role of alternative splice forms
- PMID: 16162175
- PMCID: PMC1351049
- DOI: 10.1111/j.1600-0749.2005.00249.x
MITF and cell proliferation: the role of alternative splice forms
Abstract
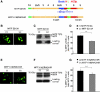
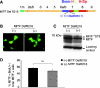
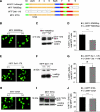
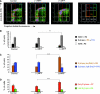
Recent studies show that the melanocyte transcription factor MITF not only activates differentiation genes but also genes involved in the regulation of the cell cycle, suggesting that it provides a link between cell proliferation and differentiation. MITF, however, comes in a variety of splice isoforms with potentially distinct biological activities. In particular, there are two isoforms, (-) and (+) MITF, that differ in six residues located upstream of the DNA binding basic domain and show slight differences in the efficiency with which they bind to target DNA. Using in vitro BrdU incorporation assays and FACS analysis in transiently transfected cells, we show that (+) MITF has a strong inhibitory effect on DNA synthesis while (-) MITF has none or only a mild one. The strong inhibitory activity of (+) MITF is not influenced by a number of mutations that modulate MITF's transcriptional activities and is independent of the protein's carboxyl terminus but dependent on its aminoterminus. A further dissection of the molecule points to the importance of an aminoterminal serine, serine-73, which in both isoforms is phosphorylated to comparable degrees. The results suggest that one or several aminoterminal domains cooperate with the alternatively spliced hexapeptide to render MITF anti-proliferative in a way that does not depend on direct E box binding.
Figures
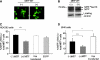
References
-
- Arnheiter H, Hou L, Nguyen M-TT, Bismuth K, Csermely T, Murakami H, Skuntz S, Liu W, Bharti K.Mitf - A Matter of Life and Death for the Developing Melanocytes Humana Press; Totowa, NJ: in press
-
- Carreira S, Liu B, Goding CR. The gene encoding the T-box factor Tbx2 is a target for the microphthalmia-associated transcription factor in melanocytes. J. Biol. Chem. 2000;275:21920–21927. - PubMed
-
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. - PubMed
-
- Del Bene F, Tessmar-Raible K, Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–749. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

