Bim is a direct target of a neuronal E2F-dependent apoptotic pathway
- PMID: 16162916
- PMCID: PMC6725681
- DOI: 10.1523/JNEUROSCI.1570-05.2005
Bim is a direct target of a neuronal E2F-dependent apoptotic pathway
Abstract
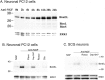
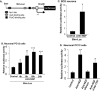
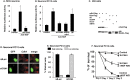
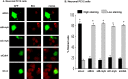
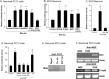
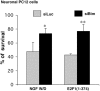
The inappropriate expression/activation of cell-cycle-related molecules is associated with neuron death in many experimental paradigms and human neuropathologic conditions. However, the means whereby this links to the core apoptotic machinery in neurons have been unclear. Here, we show that the pro-apoptotic Bcl-2 homology 3 domain-only molecule Bcl-2 interacting mediator of cell death (Bim) is a target of a cell-cycle-related apoptotic pathway in neuronal cells. Induction of Bim in NGF-deprived cells requires expression and activity of cyclin-dependent kinase 4 (cdk4) and consequent de-repression of E2 promoter binding factor (E2F)-regulated genes including members of the myb transcription factor family. The Bim promoter contains two myb binding sites, mutation of which abolishes induction of a Bim promoter-driven reporter by NGF deprivation or E2F-dependent gene de-repression. NGF deprivation significantly increases endogenous levels of C-myb and its occupancy of the endogenous Bim promoter. These findings support a model in which apoptotic stimuli lead to cdk4 activation, consequent de-repression of E2F-regulated mybs, and induction of pro-apoptotic Bim.
Figures


References
-
- Biswas SC, Greene LA (2002) Nerve growth factor (NGF) down-regulates the Bcl-2 homology 3 (BH3) domain-only protein Bim and suppresses its proapoptotic activity by phosphorylation. J Biol Chem 277: 49511–49516. - PubMed
-
- Bouillet P, Zhang LC, Huang DC, Webb GC, Bottema CD, Shore P, Eyre HJ, Sutherland GR, Adams JM (2001) Gene structure alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mamm Genome 12: 163–168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
