Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis
- PMID: 16166256
- PMCID: PMC1203353
- DOI: 10.1104/pp.105.063743
Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis
Abstract
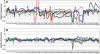
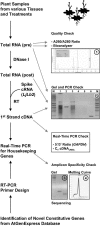
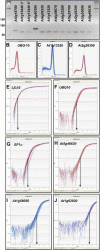
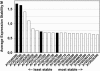
Gene transcripts with invariant abundance during development and in the face of environmental stimuli are essential reference points for accurate gene expression analyses, such as RNA gel-blot analysis or quantitative reverse transcription-polymerase chain reaction (PCR). An exceptionally large set of data from Affymetrix ATH1 whole-genome GeneChip studies provided the means to identify a new generation of reference genes with very stable expression levels in the model plant species Arabidopsis (Arabidopsis thaliana). Hundreds of Arabidopsis genes were found that outperform traditional reference genes in terms of expression stability throughout development and under a range of environmental conditions. Most of these were expressed at much lower levels than traditional reference genes, making them very suitable for normalization of gene expression over a wide range of transcript levels. Specific and efficient primers were developed for 22 genes and tested on a diverse set of 20 cDNA samples. Quantitative reverse transcription-PCR confirmed superior expression stability and lower absolute expression levels for many of these genes, including genes encoding a protein phosphatase 2A subunit, a coatomer subunit, and an ubiquitin-conjugating enzyme. The developed PCR primers or hybridization probes for the novel reference genes will enable better normalization and quantification of transcript levels in Arabidopsis in the future.
Figures

References
-
- Altmann T, Weigel D, Nover L (2004) AtGenExpress—Ein multinational koordiniertes Programm zur Erforschung des Arabidopsis Transkriptoms. GenomXpress 3: 13–14
-
- Andersen CL, Jensen JK, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250 - PubMed
-
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19: 185–193 - PubMed
-
- Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR RT-PCR: trends and problems. J Mol Endocrinol 29: 23–29 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

