Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis
- PMID: 16166381
- PMCID: PMC1221890
- DOI: 10.1101/gad.351605
Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis
Abstract
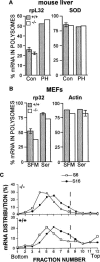
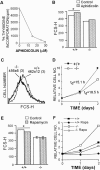
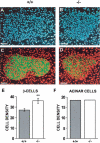
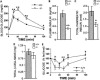
The regulated phosphorylation of ribosomal protein (rp) S6 has attracted much attention since its discovery in 1974, yet its physiological role has remained obscure. To directly address this issue, we have established viable and fertile knock-in mice, whose rpS6 contains alanine substitutions at all five phosphorylatable serine residues (rpS6(P-/-)). Here we show that contrary to the widely accepted model, this mutation does not affect the translational control of TOP mRNAs. rpS6(P-/-) mouse embryo fibroblasts (MEFs) display an increased rate of protein synthesis and accelerated cell division, and they are significantly smaller than rpS6(P+/+) MEFs. This small size reflects a growth defect, rather than a by-product of their faster cell division. Moreover, the size of rpS6(P-/-) MEFs, unlike wild-type MEFs, is not further decreased upon rapamycin treatment, implying that the rpS6 is a critical downstream effector of mTOR in regulation of cell size. The small cell phenotype is not confined to embryonal cells, as it also selectively characterizes pancreatic beta-cells in adult rpS6(P-/-) mice. These mice suffer from diminished levels of pancreatic insulin, hypoinsulinemia, and impaired glucose tolerance.
Figures



References
-
- Burgering B.M.T. and Coffer, P.J. 1995. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376: 599-602. - PubMed
-
- Chan Y. and Wool, I. 1988. The primary structure of rat ribosomal protein S6. J. Biol. Chem. 263: 2891-2896. - PubMed
-
- Chung J., Kuo, C.J., Crabtree, G.R., and Blenis, J. 1992. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kD S6 kinases. Cell 69: 1227-1236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
