A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study
- PMID: 16166543
- PMCID: PMC1251581
- DOI: 10.1128/JB.187.19.6804-6815.2005
A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study
Abstract
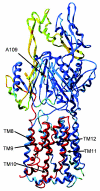
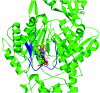
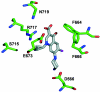
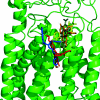
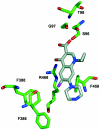
The Escherichia coli AcrB multidrug efflux pump is a membrane protein that recognizes many structurally dissimilar toxic compounds. We previously reported the X-ray structures of four AcrB-ligand complexes in which the ligands were bound to the wall of the extremely large central cavity in the transmembrane domain of the pump. Genetic studies, however, suggested that discrimination between the substrates occurs mainly in the periplasmic domain rather than the transmembrane domain of the pump. We here describe the crystal structures of the AcrB mutant in which Asn109 was replaced by Ala, with five structurally diverse ligands, ethidium, rhodamine 6G, ciprofloxacin, nafcillin, and Phe-Arg-beta-naphthylamide. The ligands bind not only to the wall of central cavity but also to a new periplasmic site within the deep external depression formed by the C-terminal periplasmic loop. This depression also includes residues identified earlier as being important in the specificity. We show here that conversion into alanine of the Phe664, Phe666, or Glu673 residue in the periplasmic binding site produced significant decreases in the MIC of most agents in the N109A background. Furthermore, decreased MICs were also observed when these residues were mutated in the wild-type AcrB background, although the effects were more modest. The MIC data were also confirmed by assays of ethidium influx rates in intact cells, and our results suggest that the periplasmic binding site plays a role in the physiological process of drug efflux.
Figures




References
-
- Akama, H., T. Matsuura, S. Kashiwagi, H. Yoneyama, T. Tsukihara, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 279:25939-25942. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Brünger, A. T. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-474. - PubMed
-
- Davidson, A. L., and H. Nikaido. 1991. Purification and characterization of the membrane-associated components of the maltose transport system from Escherichia coli. J. Biol. Chem. 266:8946-8951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

