The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion
- PMID: 16166639
- PMCID: PMC1265732
- DOI: 10.1128/MCB.25.19.8581-8591.2005
The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion
Abstract
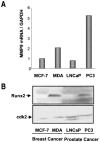
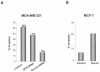
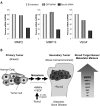
The Runx2 (Cbfa1/AML3) transcription factor and matrix metalloproteinase 9 (MMP9) are key regulators of growth plate maturation and bone formation. The genes for both proteins are characteristic markers of breast and prostate cancer cells that metastasize to bone. Here we experimentally addressed the compelling question of whether Runx2 and MMP are functionally linked. By cDNA expression array analysis, we identified MMP9 as a novel downstream target of Runx2. Like that of MMP13, MMP9 expression is nearly depleted in Runx2 mutant mice. Chromatin immunoprecipitation and electrophoretic mobility shift assays revealed the recruitment of Runx2 to the MMP9 promoter. We show by mutational analysis that the Runx2 site mediates transactivation of the MMP9 promoter in osteoblasts (MC3T3-E1) and nonosseous (HeLa) cells. The overexpression of Runx2 by adenovirus delivery in nonmetastatic (MCF-7) and metastatic breast (MDA-MB-231) and prostate (PC3) cancer cell lines significantly increases the endogenous levels of MMP9. The knockdown of Runx2 by RNA interference decreases MMP9 expression, as well as that of other Runx2 target genes, including the genes for MMP13 and vascular endothelial growth factor. Importantly, we have demonstrated using a cell invasion assay that Runx2-regulated MMP9 levels are functionally related to the invasion properties of cancer cells. These results are consistent with Runx2 control of multiple genes that contribute to the metastatic properties of cancer cells and their activity in the bone microenvironment.
Figures




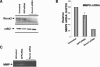
References
-
- Aalinkeel, R., M. P. Nair, G. Sufrin, S. D. Mahajan, K. C. Chadha, R. P. Chawda, and S. A. Schwartz. 2004. Gene expression of angiogenic factors correlates with metastatic potential of prostate cancer cells. Cancer Res. 64:5311-5321. - PubMed
-
- Afzal, F., J. Pratap, K. Ito, Y. Ito, J. L. Stein, A. J. van Wijnen, G. S. Stein, J. B. Lian, and A. Javed. 2005. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J. Cell. Physiol. 204:63-72. - PubMed
-
- Banerjee, C., L. R. McCabe, J.-Y. Choi, S. W. Hiebert, J. L. Stein, G. S. Stein, and J. B. Lian. 1997. Runt homology domain proteins in osteoblast differentiation: AML-3/CBFA1 is a major component of a bone specific complex. J. Cell. Biochem. 66:1-8. - PubMed
-
- Barnes, G. L., K. E. Hebert, M. Kamal, A. Javed, T. A. Einhorn, J. B. Lian, G. S. Stein, and L. C. Gerstenfeld. 2004. Fidelity of Runx2 activity in breast cancer cells is required for the generation of metastases associated osteolytic disease. Cancer Res. 64:4506-4513. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
