PubNet: a flexible system for visualizing literature derived networks
- PMID: 16168087
- PMCID: PMC1242215
- DOI: 10.1186/gb-2005-6-9-r80
PubNet: a flexible system for visualizing literature derived networks
Abstract
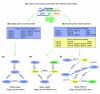
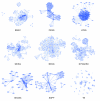
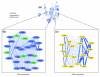
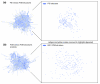
We have developed PubNet, a web-based tool that extracts several types of relationships returned by PubMed queries and maps them into networks, allowing for graphical visualization, textual navigation, and topological analysis. PubNet supports the creation of complex networks derived from the contents of individual citations, such as genes, proteins, Protein Data Bank (PDB) IDs, Medical Subject Headings (MeSH) terms, and authors. This feature allows one to, for example, examine a literature derived network of genes based on functional similarity.
Figures


References
-
- Entrez PubMed http://www.ncbi.nlm.nih.gov/entrez/
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

