The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis
- PMID: 16169895
- PMCID: PMC1242268
- DOI: 10.1105/tpc.105.034975
The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis
Abstract
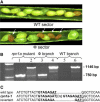
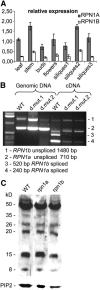
The 26S proteasome plays a central role in the degradation of regulatory proteins involved in a variety of developmental processes. It consists of two multisubunit protein complexes: the proteolytic core protease and the regulatory particle (RP). The function of most RP subunits is poorly understood. Here, we describe mutants in the Arabidopsis thaliana RPN1 subunit, which is encoded by two paralogous genes, RPN1a and RPN1b. Disruption of RPN1a caused embryo lethality, while RPN1b mutants showed no obvious abnormal phenotype. Embryos homozygous for rpn1a arrested at the globular stage with defects in the formation of the embryonic root, the protoderm, and procambium. Cyclin B1 protein was not degraded in these embryos, consistent with cell division defects. Double mutant plants (rpn1a/RPN1a rpn1b/rpn1b) produced embryos with a phenotype indistinguishable from that of the rpn1a single mutant. Thus, despite their largely overlapping expression patterns in flowers and developing seeds, the two isoforms do not share redundant functions during gametogenesis and embryogenesis. However, complementation of the rpn1a mutation with the coding region of RPN1b expressed under the control of the RPN1a promoter indicates that the two RPN1 isoforms are functionally equivalent. Overall, our data indicate that RPN1 activity is essential during embryogenesis, where it might participate in the destruction of a specific set of protein substrates.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

