Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis
- PMID: 16169902
- PMCID: PMC1236569
- DOI: 10.1073/pnas.0506447102
Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis
Abstract
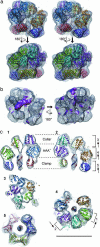
Ring-shaped sliding clamps and clamp loader ATPases are essential factors for rapid and accurate DNA replication. The clamp ring is opened and resealed at the primer-template junctions by the ATP-fueled clamp loader function. The processivity of the DNA polymerase is conferred by its attachment to the clamp loaded onto the DNA. In eukarya and archaea, the replication factor C (RFC) and the proliferating cell nuclear antigen (PCNA) play crucial roles as the clamp loader and the clamp, respectively. Here, we report the electron microscopic structure of an archaeal RFC-PCNA-DNA complex at 12-A resolution. This complex exhibits excellent fitting of each atomic structure of RFC, PCNA, and the primed DNA. The PCNA ring retains an open conformation by extensive interactions with RFC, with a distorted spring washer-like conformation. The complex appears to represent the intermediate, where the PCNA ring is kept open before ATP hydrolysis by RFC.
Figures



Comment in
-
The opened processivity clamp slides into view.Proc Natl Acad Sci U S A. 2005 Oct 18;102(42):14939-40. doi: 10.1073/pnas.0507120102. Epub 2005 Oct 10. Proc Natl Acad Sci U S A. 2005. PMID: 16217036 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

