Sex chromosome evolution: molecular aspects of Y-chromosome degeneration in Drosophila
- PMID: 16169921
- PMCID: PMC1240082
- DOI: 10.1101/gr.3543605
Sex chromosome evolution: molecular aspects of Y-chromosome degeneration in Drosophila
Abstract
Ancient Y-chromosomes of various organisms contain few active genes and an abundance of repetitive DNA. The neo-Y chromosome of Drosophila miranda is in transition from an ordinary autosome to a genetically inert Y-chromosome, while its homolog, the neo-X chromosome, is evolving partial dosage compensation. Here, I compare four large genomic regions located on the neo-sex chromosomes that contain a total of 12 homologous genes. In addition, I investigate the partial coding sequence for 56 more homologous gene pairs from the neo-sex chromosomes. Little modification has occurred on the neo-X chromosome, and genes are highly constrained at the protein level. In contrast, a diverse array of molecular changes is contributing to the observed degeneration of the neo-Y chromosome. In particular, the four large regions surveyed on the neo-Y chromosome harbor several transposable element insertions, large deletions, and a large structural rearrangement. About one-third of all neo-Y-linked genes are nonfunctional, containing either premature stop codons and/or frameshift mutations. Intact genes on the neo-Y are accumulating amino acid and unpreferred codon changes. In addition, both 5'- and 3'-flanking regions of genes and intron sequences are less constrained on the neo-Y relative to the neo-X. Despite heterogeneity in levels of dosage compensation along the neo-X chromosome of D. miranda, the neo-Y chromosome shows surprisingly uniform signs of degeneration.
Figures
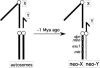


References
-
- Bachtrog, D. 2003a. Adaptation shapes patterns of genome evolution in sexual and asexual genomes in Drosophila. Nat. Genet. 34: 215-219. - PubMed
-
- ———. 2003b. Accumulation of spock and worf, two novel non-LTR retrotransposons on the neo-Y chromosome of Drosophila miranda. Mol. Biol. Evol. 20: 173-181. - PubMed
WEB SITE REFERENCES
-
- http://flybase.bio.indiana.edu/blast/; FlyBase.
-
- http://www.ebi.ac.uk/Wise2/; Wise2.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
