Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125
- PMID: 16169927
- PMCID: PMC1240074
- DOI: 10.1101/gr.4126905
Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125
Abstract
A considerable fraction of life develops in the sea at temperatures lower than 15 degrees C. Little is known about the adaptive features selected under those conditions. We present the analysis of the genome sequence of the fast growing Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. We find that it copes with the increased solubility of oxygen at low temperature by multiplying dioxygen scavenging while deleting whole pathways producing reactive oxygen species. Dioxygen-consuming lipid desaturases achieve both protection against oxygen and synthesis of lipids making the membrane fluid. A remarkable strategy for avoidance of reactive oxygen species generation is developed by P. haloplanktis, with elimination of the ubiquitous molybdopterin-dependent metabolism. The P. haloplanktis proteome reveals a concerted amino acid usage bias specific to psychrophiles, consistently appearing apt to accommodate asparagine, a residue prone to make proteins age. Adding to its originality, P. haloplanktis further differs from its marine counterparts with recruitment of a plasmid origin of replication for its second chromosome.
Figures

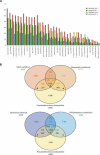

References
-
- Barber, C.E., Tang, J.L., Feng, J.X., Pan, M.Q., Wilson, T.J., Slater, H., Dow, J.M., Williams, P., and Daniels, M.J. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24: 555-566. - PubMed
-
- Bartlett, D.H. 1999. Microbial adaptations to the psychrosphere/piezosphere. J. Mol. Microbiol. Biotechnol. 1: 93-100. - PubMed
-
- Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783-795. - PubMed
-
- Benzécri, J.P. 1984. L'analyse des données. L'analyse des correspondances. Dunod, Paris.
WEB SITE REFERENCES
-
- www.genoscope.cns.fr/agc/mage/psychroscope; PsychroScope http://bioinfo.khu.hk/PsychroList; PsychroListd
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
