Determination of catalytic key amino acids and UDP sugar donor specificity of the cyanohydrin glycosyltransferase UGT85B1 from Sorghum bicolor. Molecular modeling substantiated by site-specific mutagenesis and biochemical analyses
- PMID: 16169969
- PMCID: PMC1255986
- DOI: 10.1104/pp.105.063842
Determination of catalytic key amino acids and UDP sugar donor specificity of the cyanohydrin glycosyltransferase UGT85B1 from Sorghum bicolor. Molecular modeling substantiated by site-specific mutagenesis and biochemical analyses
Abstract
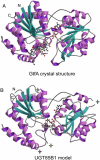
Plants produce a plethora of structurally diverse natural products. The final step in their biosynthesis is often a glycosylation step catalyzed by a family 1 glycosyltransferase (GT). In biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor, the UDP-glucosyltransferase UGT85B1 catalyzes the conversion of p-hydroxymandelonitrile into dhurrin. A structural model of UGT85B1 was built based on hydrophobic cluster analysis and the crystal structures of two bacterial GTs, GtfA and GtfB, which each showed approximately 15% overall amino acid sequence identity to UGT85B1. The model enabled predictions about amino acid residues important for catalysis and sugar donor specificity. p-Hydroxymandelonitrile and UDP-glucose (Glc) were predicted to be positioned within hydrogen-bonding distance to a glutamic acid residue in position 410 facilitating sugar transfer. The acceptor was packed within van der Waals distance to histidine H23. Serine S391 and arginine R201 form hydrogen bonds to the pyrophosphate part of UDP-Glc and hence stabilize binding of the sugar donor. Docking of UDP sugars predicted that UDP-Glc would serve as the sole donor sugar in UGT85B1. This was substantiated by biochemical analyses. The predictive power of the model was validated by site-directed mutagenesis of selected residues and using enzyme assays. The modeling approach has provided a tool to design GTs with new desired substrate specificities for use in biotechnological applications. The modeling identified a hypervariable loop (amino acid residues 156-188) that contained a hydrophobic patch. The involvement of this loop in mediating binding of UGT85B1 to cytochromes P450, CYP79A1, and CYP71E1 within a dhurrin metabolon is discussed.
Figures




Similar articles
-
Metabolic consequences of knocking out UGT85B1, the gene encoding the glucosyltransferase required for synthesis of dhurrin in Sorghum bicolor (L. Moench).Plant Cell Physiol. 2016 Feb;57(2):373-86. doi: 10.1093/pcp/pcv153. Epub 2015 Oct 22. Plant Cell Physiol. 2016. PMID: 26493517
-
Metabolon formation in dhurrin biosynthesis.Phytochemistry. 2008 Jan;69(1):88-98. doi: 10.1016/j.phytochem.2007.06.033. Epub 2007 Aug 15. Phytochemistry. 2008. PMID: 17706731
-
Homology modeling of the three membrane proteins of the dhurrin metabolon: catalytic sites, membrane surface association and protein-protein interactions.Phytochemistry. 2011 Dec;72(17):2113-23. doi: 10.1016/j.phytochem.2011.05.001. Epub 2011 May 26. Phytochemistry. 2011. PMID: 21620426
-
Insights into the UDP-sugar selectivities of human UDP-glycosyltransferases (UGT): a molecular modeling perspective.Drug Metab Rev. 2015 Aug;47(3):335-45. doi: 10.3109/03602532.2015.1071835. Epub 2015 Aug 3. Drug Metab Rev. 2015. PMID: 26289097 Review.
-
Identification of UDP-glucose binding site in glycosyltransferase domain of sucrose phosphate synthase from sugarcane (Saccharum officinarum) by structure-based site-directed mutagenesis.Biophys Rev. 2018 Apr;10(2):293-298. doi: 10.1007/s12551-017-0360-9. Epub 2017 Dec 8. Biophys Rev. 2018. PMID: 29222806 Free PMC article. Review.
Cited by
-
Molecular identification and functional characterization of a cyanogenic glucosyltransferase from flax (Linum unsitatissimum).PLoS One. 2020 Feb 5;15(2):e0227840. doi: 10.1371/journal.pone.0227840. eCollection 2020. PLoS One. 2020. PMID: 32023283 Free PMC article.
-
Selective Detoxification of Phenols by Pichia pastoris and Arabidopsis thaliana Heterologously Expressing the PtUGT72B1 from Populus trichocarpa.PLoS One. 2013 Jun 26;8(6):e66878. doi: 10.1371/journal.pone.0066878. Print 2013. PLoS One. 2013. PMID: 23840543 Free PMC article.
-
Digital gene expression analysis of male and female bud transition in Metasequoia reveals high activity of MADS-box transcription factors and hormone-mediated sugar pathways.Front Plant Sci. 2015 Jun 24;6:467. doi: 10.3389/fpls.2015.00467. eCollection 2015. Front Plant Sci. 2015. PMID: 26157452 Free PMC article.
-
Glucosylation of 4-Hydroxy-2,5-Dimethyl-3(2H)-Furanone, the Key Strawberry Flavor Compound in Strawberry Fruit.Plant Physiol. 2016 May;171(1):139-51. doi: 10.1104/pp.16.00226. Epub 2016 Mar 18. Plant Physiol. 2016. PMID: 26993618 Free PMC article.
-
UDP-glycosyltransferases from the UGT73C subfamily in Barbarea vulgaris catalyze sapogenin 3-O-glucosylation in saponin-mediated insect resistance.Plant Physiol. 2012 Dec;160(4):1881-95. doi: 10.1104/pp.112.202747. Epub 2012 Oct 1. Plant Physiol. 2012. PMID: 23027665 Free PMC article.
References
-
- Blundell T, Carney D, Gardner S, Hayes F, Howlin B, Hubbard T, Overington J, Singh DA, Sibanda BL, Sutcliffe M (1988) 18th Sir Hans Krebs lecture. Knowledge-based protein modelling and design. Eur J Biochem 172: 513–520 - PubMed
-
- Breton C, Heissigerova H, Jeanneau C, Moravcova J, Imberty A (2002) Comparative aspects of glycosyltransferases. Biochem Soc Symp 69: 23–32 - PubMed
-
- Clark M, Cramer RDI, van den Opdenbosch N (1989) Validation of the general purpose Tripos 5.2 force field. J Comput Chem 10: 982–1012
-
- Combet C, Blanchet C, Geourjon C, Deleage G (2000) NPS@: network protein sequence analysis. Trends Biochem Sci 25: 147–150 - PubMed
-
- Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

