The conformational stability and thermodynamics of Fur A (ferric uptake regulator) from Anabaena sp. PCC 7119
- PMID: 16169981
- PMCID: PMC1366984
- DOI: 10.1529/biophysj.105.065805
The conformational stability and thermodynamics of Fur A (ferric uptake regulator) from Anabaena sp. PCC 7119
Abstract
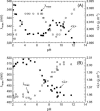
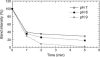
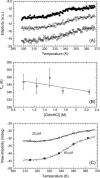
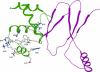
Fur (ferric uptake regulator) is a key bacterial protein that regulates iron acquisition and its storage, and modulates the expression of genes involved in the response to different environmental stresses. Although the protein is involved in several regulation mechanisms, and members of the Fur family have been identified in pathogen organisms, the stability and thermodynamic characterization of a Fur protein have not been described. In this work, the stability, thermodynamics and structure of the functional dimeric Fur A from Anabaena sp. PCC 7119 were studied by using computational methods and different biophysical techniques, namely, circular dichroism, fluorescence, Fourier-transform infrared, and nuclear magnetic resonance spectroscopies. The structure, as monitored by circular dichroism and Fourier-transform infrared, was composed of a 40% of alpha-helix. Chemical-denaturation experiments indicated that Fur A folded via a two-state mechanism, but its conformational stability was small with a value of DeltaG = 5.3 +/- 0.3 kcal mol(-1) at 298 K. Conversely, Fur A was thermally a highly stable protein. The high melting temperature (Tm = 352 +/- 5 K), despite its moderate conformational stability, can be ascribed to its low heat capacity change upon unfolding, DeltaCp, which had a value of 0.8 +/- 0.1 kcal mol(-1) K(-1). This small value is probably due to burial of polar residues in the Fur A structure. This feature can be used for the design of mutants of Fur A with impaired DNA-binding properties.
Figures




References
-
- Braun, V., and H. Killmann. 1999. Bacterial solutions to the iron supply problem. Trends Biochem. Sci. 24:104–109. - PubMed
-
- Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881–941. - PubMed
-
- Escolar, L., V. de Lorenzo, and J. Pérez-Martín. 1997. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulator) protein. Mol. Microbiol. 26:799–808. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

