The hnRNPs F and H2 bind to similar sequences to influence gene expression
- PMID: 16171461
- PMCID: PMC1383695
- DOI: 10.1042/BJ20050538
The hnRNPs F and H2 bind to similar sequences to influence gene expression
Abstract
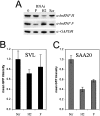
The hnRNPs (heterogeneous nuclear ribonucleoproteins) F and H2 share a similar protein structure. Both have been implicated as regulating polyadenylation, but hnRNP H2 had a positive effect, whereas hnRNP F acted negatively. We therefore carried out side-by-side comparisons of their RNA-binding and in vivo actions. The binding of the CstF2 (64 kDa cleavage stimulatory factor) to SV40 (simian virus 40) late pre-mRNA substrates containing a downstream GRS (guanine-rich sequence) was reduced by hnRNP F, but not by hnRNP H2, in a UV-cross-linking assay. Point mutations of the 14-nt GRS influenced the binding of purified hnRNP F or H2 in parallel. Co-operative binding of the individual proteins to RNA was lost with mutations of the GRS in the G1-5 or G12-14 regions; both regions seem to be necessary for optimal interactions. Using a reporter green fluorescent protein assay with the GRS inserted downstream of the poly(A) (polyadenine) signal, expression in vivo was diminished by a mutant G1-5 sequence which decreased binding of both hnRNPs (SAA20) and was enhanced by a 12-14-nt mutant that showed enhanced hnRNP F or H2 binding (SAA10). Using small interfering RNA, down-regulation of hnRNP H2 levels diminished reporter expression, confirming that hnRNP H2 confers a positive influence; in contrast, decreasing hnRNP F levels had a negligible influence on reporter expression with the intact GRS. A pronounced diminution in reporter expression was seen with the SAA20 mutant for both. Thus the relative levels of hnRNP F and H2 in cells, as well as the target sequences in the downstream GRS on pre-mRNA, influence gene expression.
Figures







Similar articles
-
hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells.Mol Cell Biol. 2001 Feb;21(4):1228-38. doi: 10.1128/MCB.21.4.1228-1238.2001. Mol Cell Biol. 2001. PMID: 11158309 Free PMC article.
-
The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization.Nucleic Acids Res. 1994 Mar 25;22(6):1059-67. doi: 10.1093/nar/22.6.1059. Nucleic Acids Res. 1994. PMID: 7512260 Free PMC article.
-
Heterogeneous nuclear ribonucleoproteins F and H/H' show differential expression in normal and selected cancer tissues.Exp Cell Res. 2004 Mar 10;294(1):199-209. doi: 10.1016/j.yexcr.2003.11.011. Exp Cell Res. 2004. PMID: 14980514
-
HnRNP-F regulates EMT in bladder cancer by mediating the stabilization of Snail1 mRNA by binding to its 3' UTR.EBioMedicine. 2019 Jul;45:208-219. doi: 10.1016/j.ebiom.2019.06.017. Epub 2019 Jun 18. EBioMedicine. 2019. PMID: 31221586 Free PMC article.
-
Regulation of HNRNP family by post-translational modifications in cancer.Cell Death Discov. 2024 Oct 4;10(1):427. doi: 10.1038/s41420-024-02198-7. Cell Death Discov. 2024. PMID: 39366930 Free PMC article. Review.
Cited by
-
p53 promotes the fidelity of DNA end-joining activity by, in part, enhancing the expression of heterogeneous nuclear ribonucleoprotein G.DNA Repair (Amst). 2007 Jun 1;6(6):830-40. doi: 10.1016/j.dnarep.2007.01.013. Epub 2007 Mar 26. DNA Repair (Amst). 2007. PMID: 17387044 Free PMC article.
-
A nonsense exon in the Tpm1 gene is silenced by hnRNP H and F.RNA. 2009 Jan;15(1):33-43. doi: 10.1261/rna.1225209. Epub 2008 Nov 26. RNA. 2009. PMID: 19037011 Free PMC article.
-
Regulation of global and specific mRNA translation by the mTOR signaling pathway.Translation (Austin). 2015 Feb 2;3(1):e983402. doi: 10.4161/21690731.2014.983402. eCollection 2015 Jan-Jun. Translation (Austin). 2015. PMID: 26779414 Free PMC article. Review.
-
Competitive regulation of alternative splicing and alternative polyadenylation by hnRNP H and CstF64 determines acetylcholinesterase isoforms.Nucleic Acids Res. 2017 Feb 17;45(3):1455-1468. doi: 10.1093/nar/gkw823. Nucleic Acids Res. 2017. PMID: 28180311 Free PMC article.
-
Splice site strength-dependent activity and genetic buffering by poly-G runs.Nat Struct Mol Biol. 2009 Oct;16(10):1094-100. doi: 10.1038/nsmb.1661. Epub 2009 Sep 13. Nat Struct Mol Biol. 2009. PMID: 19749754 Free PMC article.
References
-
- Krecic A. M., Swanson M. S. HnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. - PubMed
-
- Rasmussen H. B., Vorum H. H., Dejgaard K., Liu X., Gromov P., Madsen P., Gesser B., Tommerup N., Celis J. E. Heterogeneous nuclear ribonucleoproteins (hnRNP) H, H′, and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem. 1995;270:28780–28789. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

