Genome annotation of Anopheles gambiae using mass spectrometry-derived data
- PMID: 16171517
- PMCID: PMC1249570
- DOI: 10.1186/1471-2164-6-128
Genome annotation of Anopheles gambiae using mass spectrometry-derived data
Abstract
Background: A large number of animal and plant genomes have been completely sequenced over the last decade and are now publicly available. Although genomes can be rapidly sequenced, identifying protein-coding genes still remains a problematic task. Availability of protein sequence data allows direct confirmation of protein-coding genes. Mass spectrometry has recently emerged as a powerful tool for proteomic studies. Protein identification using mass spectrometry is usually carried out by searching against databases of known proteins or transcripts. This approach generally does not allow identification of proteins that have not yet been predicted or whose transcripts have not been identified.
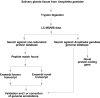
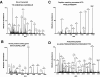
Results: We searched 3,967 mass spectra from 16 LC-MS/MS runs of Anopheles gambiae salivary gland homogenates against the Anopheles gambiae genome database. This allowed us to validate 23 known transcripts and 50 novel transcripts. In addition, a novel gene was identified on the basis of peptides that matched a genomic region where no gene was known and no transcript had been predicted. The amino termini of proteins encoded by two predicted transcripts were confirmed based on N-terminally acetylated peptides sequenced by tandem mass spectrometry. Finally, six sequence polymorphisms could be annotated based on experimentally obtained peptide sequences.
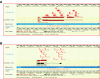
Conclusion: The peptide sequences from this study were mapped onto the genomic sequence using the distributed annotation system available at Ensembl and can be visualized in the context of all other existing annotations. The strategy described in this paper can be used to correct and confirm genome annotations and permit discovery of novel proteins in a high-throughput manner by mass spectrometry.
Figures




Similar articles
-
Proteome research: complementarity and limitations with respect to the RNA and DNA worlds.Electrophoresis. 1997 Aug;18(8):1217-42. doi: 10.1002/elps.1150180804. Electrophoresis. 1997. PMID: 9298643 Review.
-
MitoRes: a resource of nuclear-encoded mitochondrial genes and their products in Metazoa.BMC Bioinformatics. 2006 Jan 24;7:36. doi: 10.1186/1471-2105-7-36. BMC Bioinformatics. 2006. PMID: 16433928 Free PMC article.
-
A proteomic analysis of salivary glands of female Anopheles gambiae mosquito.Proteomics. 2005 Sep;5(14):3765-77. doi: 10.1002/pmic.200401210. Proteomics. 2005. PMID: 16127729
-
VEMS 3.0: algorithms and computational tools for tandem mass spectrometry based identification of post-translational modifications in proteins.J Proteome Res. 2005 Nov-Dec;4(6):2338-47. doi: 10.1021/pr050264q. J Proteome Res. 2005. PMID: 16335983
-
Comprehensive mass spectrometric analysis of the 20S proteasome complex.Methods Enzymol. 2005;405:187-236. doi: 10.1016/S0076-6879(05)05009-3. Methods Enzymol. 2005. PMID: 16413316 Review.
Cited by
-
Comparative Proteogenomic Approaches for Mapping the Global Proteome of the Unsequenced Leishmania Vector Phlebotomus papatasi.Methods Mol Biol. 2025;2859:265-277. doi: 10.1007/978-1-0716-4152-1_15. Methods Mol Biol. 2025. PMID: 39436607
-
Addressing statistical biases in nucleotide-derived protein databases for proteogenomic search strategies.J Proteome Res. 2012 Nov 2;11(11):5221-34. doi: 10.1021/pr300411q. Epub 2012 Oct 15. J Proteome Res. 2012. PMID: 23025403 Free PMC article.
-
Moving from unsequenced to sequenced genome: reanalysis of the proteome of Leishmania donovani.J Proteomics. 2014 Jan 31;97:48-61. doi: 10.1016/j.jprot.2013.04.021. Epub 2013 May 9. J Proteomics. 2014. PMID: 23665000 Free PMC article.
-
Proteomic profiling of the planarian Schmidtea mediterranea and its mucous reveals similarities with human secretions and those predicted for parasitic flatworms.Mol Cell Proteomics. 2012 Sep;11(9):681-91. doi: 10.1074/mcp.M112.019026. Epub 2012 May 31. Mol Cell Proteomics. 2012. PMID: 22653920 Free PMC article.
-
Accelerating string set matching in FPGA hardware for bioinformatics research.BMC Bioinformatics. 2008 Apr 15;9:197. doi: 10.1186/1471-2105-9-197. BMC Bioinformatics. 2008. PMID: 18412963 Free PMC article.
References
-
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O'Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. - DOI - PubMed
-
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci U S A. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

