Diversity in domain architectures of Ser/Thr kinases and their homologues in prokaryotes
- PMID: 16171520
- PMCID: PMC1262709
- DOI: 10.1186/1471-2164-6-129
Diversity in domain architectures of Ser/Thr kinases and their homologues in prokaryotes
Abstract
Background: Ser/Thr/Tyr kinases (STYKs) commonly found in eukaryotes have been recently reported in many bacterial species. Recent studies elucidating their cellular functions have established their roles in bacterial growth and development. However functions of a large number of bacterial STYKs still remain elusive. The organisation of domains in a large dataset of bacterial STYKs has been investigated here in order to recognise variety in domain combinations which determine functions of bacterial STYKs.
Results: Using sensitive sequence and profile search methods, domain organisation of over 600 STYKs from 125 prokaryotic genomes have been examined. Kinase catalytic domains of STYKs tethered to a wide range of enzymatic domains such as phosphatases, HSP70, peptidyl prolyl isomerases, pectin esterases and glycoproteases have been identified. Such distinct preferences for domain combinations are not known to be present in either the Histidine kinase or the eukaryotic STYK families. Domain organisation of STYKs specific to certain groups of bacteria has also been noted in the current anlaysis. For example, Hydrophobin like domains in Mycobacterial STYK and penicillin binding domains in few STYKs of Gram-positive organisms and FHA domains in cyanobacterial STYKs. Homologues of characterised substrates of prokaryotic STYKs have also been identified.
Conclusion: The domains and domain architectures of most of the bacterial STYKs identified are very different from the known domain organisation in STYKs of eukaryotes. This observation highlights distinct biological roles of bacterial STYKs compared to eukaryotic STYKs. Bacterial STYKs reveal high diversity in domain organisation. Some of the modular organisations conserved across diverse bacterial species suggests their central role in bacterial physiology. Unique domain architectures of few other groups of STYKs reveal recruitment of functions specific to the species.
Figures

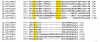

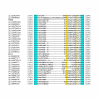
Similar articles
-
Eukaryote-like serine/threonine kinases and phosphatases in bacteria.Microbiol Mol Biol Rev. 2011 Mar;75(1):192-212. doi: 10.1128/MMBR.00042-10. Microbiol Mol Biol Rev. 2011. PMID: 21372323 Free PMC article. Review.
-
A large family of eukaryotic-like protein Ser/Thr kinases of Myxococcus xanthus, a developmental bacterium.Microb Comp Genomics. 2000;5(2):103-20. doi: 10.1089/10906590050179783. Microb Comp Genomics. 2000. PMID: 11087177
-
Evolution of domain combinations in protein kinases and its implications for functional diversity.Prog Biophys Mol Biol. 2010 Jan;102(1):1-15. doi: 10.1016/j.pbiomolbio.2009.12.009. Epub 2009 Dec 22. Prog Biophys Mol Biol. 2010. PMID: 20026163 Review.
-
Genome-wide survey of putative serine/threonine protein kinases in cyanobacteria.BMC Genomics. 2007 Oct 30;8:395. doi: 10.1186/1471-2164-8-395. BMC Genomics. 2007. PMID: 17971218 Free PMC article.
-
HstK, a cyanobacterial protein with both a serine/threonine kinase domain and a histidine kinase domain: implication for the mechanism of signal transduction.Biochem J. 2001 Dec 15;360(Pt 3):639-44. doi: 10.1042/0264-6021:3600639. Biochem J. 2001. PMID: 11736654 Free PMC article.
Cited by
-
Phylogeny and Expansion of Serine/Threonine Kinases in Phagocytotic Bacteria in the Phylum Planctomycetota.Genome Biol Evol. 2024 Apr 2;16(4):evae068. doi: 10.1093/gbe/evae068. Genome Biol Evol. 2024. PMID: 38547507 Free PMC article.
-
Tracing the origin and evolution of pseudokinases across the tree of life.Sci Signal. 2019 Apr 23;12(578):eaav3810. doi: 10.1126/scisignal.aav3810. Sci Signal. 2019. PMID: 31015289 Free PMC article.
-
Revisiting cyanobacterial state transitions.Photochem Photobiol Sci. 2020 May 1;19(5):585-603. doi: 10.1039/c9pp00451c. Epub 2020 Mar 12. Photochem Photobiol Sci. 2020. PMID: 32163064 Review.
-
Non-Receptor Tyrosine Kinases: Their Structure and Mechanistic Role in Tumor Progression and Resistance.Cancers (Basel). 2024 Aug 2;16(15):2754. doi: 10.3390/cancers16152754. Cancers (Basel). 2024. PMID: 39123481 Free PMC article. Review.
-
A framework for classification of prokaryotic protein kinases.PLoS One. 2010 May 26;5(5):e10608. doi: 10.1371/journal.pone.0010608. PLoS One. 2010. PMID: 20520783 Free PMC article.
References
-
- Hurley JH, Dean AM, Thorsness PE, Koshland DE, Jr, Stroud RM. Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the free enzyme. J Biol Chem. 1990;265:3599–3602. - PubMed
-
- Hunter T. Protein kinase classification. Methods Enzymol. 1991;200:3–37. - PubMed
-
- Duclos B, Marcandier S, Cozzone AJ. Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol. 1991;201:10–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

