Chromosomal clustering of a human transcriptome reveals regulatory background
- PMID: 16171528
- PMCID: PMC1261156
- DOI: 10.1186/1471-2105-6-230
Chromosomal clustering of a human transcriptome reveals regulatory background
Abstract
Background: There has been much evidence recently for a link between transcriptional regulation and chromosomal gene order, but the relationship between genomic organization, regulation and gene function in higher eukaryotes remains to be precisely defined.
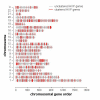
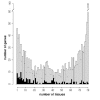
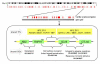
Results: Here, we present evidence for organization of a large proportion of a human transcriptome into gene clusters throughout the genome, which are partly regulated by the same transcription factors, share biological functions and are characterized by non-housekeeping genes. This analysis was based on the cardiac transcriptome identified by our genome-wide array analysis of 55 human heart samples. We found 37% of these genes to be arranged mainly in adjacent pairs or triplets. A significant number of pairs of adjacent genes are putatively regulated by common transcription factors (p = 0.02). Furthermore, these gene pairs share a significant number of GO functional classification terms. We show that the human cardiac transcriptome is organized into many small clusters across the whole genome, rather than being concentrated in a few larger clusters.
Conclusion: Our findings suggest that genes expressed in concert are organized in a linear arrangement for coordinated regulation. Determining the relationship between gene arrangement, regulation and nuclear organization as well as gene function will have broad biological implications.
Figures
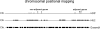
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

