Dragon Plant Biology Explorer. A text-mining tool for integrating associations between genetic and biochemical entities with genome annotation and biochemical terms lists
- PMID: 16172098
- PMCID: PMC1183383
- DOI: 10.1104/pp.105.060863
Dragon Plant Biology Explorer. A text-mining tool for integrating associations between genetic and biochemical entities with genome annotation and biochemical terms lists
Abstract
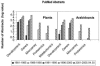
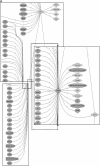
We introduce a tool for text mining, Dragon Plant Biology Explorer (DPBE) that integrates information on Arabidopsis (Arabidopsis thaliana) genes with their functions, based on gene ontologies and biochemical entity vocabularies, and presents the associations as interactive networks. The associations are based on (1) user-provided PubMed abstracts; (2) a list of Arabidopsis genes compiled by The Arabidopsis Information Resource; (3) user-defined combinations of four vocabulary lists based on the ones developed by the general, plant, and Arabidopsis GO consortia; and (4) three lists developed here based on metabolic pathways, enzymes, and metabolites derived from AraCyc, BRENDA, and other metabolism databases. We demonstrate how various combinations can be applied to fields of (1) gene function and gene interaction analyses, (2) plant development, (3) biochemistry and metabolism, and (4) pharmacology of bioactive compounds. Furthermore, we show the suitability of DPBE for systems approaches by integration with "omics" platform outputs. Using a list of abiotic stress-related genes identified by microarray experiments, we show how this tool can be used to rapidly build an information base on the previously reported relationships. This tool complements the existing biological resources for systems biology by identifying potentially novel associations using text analysis between cellular entities based on genome annotation terms. Thus, it allows researchers to efficiently summarize existing information for a group of genes or pathways, so as to make better informed choices for designing validation experiments. Last, DPBE can be helpful for beginning researchers and graduate students to summarize vast information in an unfamiliar area. DPBE is freely available for academic and nonprofit users at http://research.i2r.a-star.edu.sg/DRAGON/ME2/.
Figures



References
-
- Andrade MA, Bork P (2000) Automated extraction of information in molecular biology. FEBS Lett 476: 12–17 - PubMed
-
- Andrade MA, Valencia A (1998) Automatic extraction of keywords from scientific knowledge: application to the knowledge domain of protein families. Bioinformatics 14: 600–607 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

