Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome"
- PMID: 16172379
- PMCID: PMC1216834
- DOI: 10.1073/pnas.0506758102
Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome"
Erratum in
- Proc Natl Acad Sci U S A. 2005 Nov 8;102(45):16530
Abstract
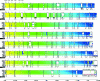
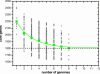
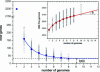
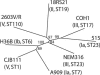
The development of efficient and inexpensive genome sequencing methods has revolutionized the study of human bacterial pathogens and improved vaccine design. Unfortunately, the sequence of a single genome does not reflect how genetic variability drives pathogenesis within a bacterial species and also limits genome-wide screens for vaccine candidates or for antimicrobial targets. We have generated the genomic sequence of six strains representing the five major disease-causing serotypes of Streptococcus agalactiae, the main cause of neonatal infection in humans. Analysis of these genomes and those available in databases showed that the S. agalactiae species can be described by a pan-genome consisting of a core genome shared by all isolates, accounting for approximately 80% of any single genome, plus a dispensable genome consisting of partially shared and strain-specific genes. Mathematical extrapolation of the data suggests that the gene reservoir available for inclusion in the S. agalactiae pan-genome is vast and that unique genes will continue to be identified even after sequencing hundreds of genomes.
Figures
References
-
- Wayne, L., Brenner, D., Colwell, R., Grimont, P., Kandler, O., Krichevsky, L., Moore, L., Moore, W., Murray, R., Stackebrandt, E., et al. (1987) Int. J. Syst. Bacteriol. 37, 463–464.
-
- Schuchat, A. & Wenger, J. D. (1994) Epidemiol. Rev. 16, 374–402. - PubMed
-
- Tyrrell, G. J., Senzilet, L. D., Spika, J. S., Kertesz, D. A., Alagaratnam, M., Lovgren, M. & Talbot, J. A. (2000) J. Infect. Dis. 182, 168–173. - PubMed
-
- Harrison, L. H., Elliott, J. A., Dwyer, D. M., Libonati, J. P., Ferrieri, P., Billmann, L. & Schuchat, A. (1998) J. Infect. Dis. 177, 998–1002. - PubMed
-
- Lin, F. Y., Clemens, J. D., Azimi, P. H., Regan, J. A., Weisman, L. E., Philips, J. B., III, Rhoads, G. G., Clark, P., Brenner, R. A. & Ferrieri, P. (1998) J. Infect. Dis. 177, 790–792. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

