Identification of slow correlated motions in proteins using residual dipolar and hydrogen-bond scalar couplings
- PMID: 16172390
- PMCID: PMC1236556
- DOI: 10.1073/pnas.0505129102
Identification of slow correlated motions in proteins using residual dipolar and hydrogen-bond scalar couplings
Abstract
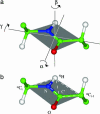
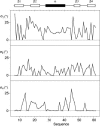
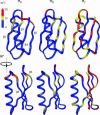
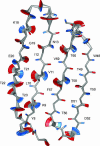
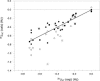
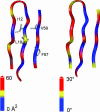
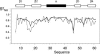
Despite their importance for biological activity, slower molecular motions beyond the nanosecond range remain poorly understood. We have assembled an unprecedented set of experimental NMR data, comprising up to 27 residual dipolar couplings per amino acid, to define the nature and amplitude of backbone motion in protein G using the Gaussian axial fluctuation model in three dimensions. Slower motions occur in the loops, and in the beta-sheet, and are absent in other regions of the molecule, including the alpha-helix. In the beta-sheet an alternating pattern of dynamics along the peptide sequence is found to form a long-range network of slow motion in the form of a standing wave extending across the beta-sheet, resulting in maximal conformational sampling at the interaction site. The alternating nodes along the sequence match the alternation of strongly hydrophobic side chains buried in the protein core. Confirmation of the motion is provided through extensive cross-validation and by independent hydrogen-bond scalar coupling analysis that shows this motion to be correlated. These observations strongly suggest that dynamical information can be transmitted across hydrogen bonds and have important implications for understanding collective motions and long-range information transfer in proteins.
Figures
References
-
- Frauenfelder, H., Sligar, S. G. & Wolynes, P. G. (1991) Science 254, 1598–1603. - PubMed
-
- Karplus, M. & Kuriyan, J. (2005) Proc. Natl. Acad. Sci. USA 9, 646–652.
-
- Eisenmesser, E. Z., Bosco, D. A., Akke, M. & Kern, D. (2002) Science 295, 1520–1523. - PubMed
-
- Benkovic, S. J. & Hammes-Schiffer, S. (2003) Science 301, 1196–1202. - PubMed
-
- Baker, D. & Sali, A. (2001) Science 294, 93–96. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

