Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation
- PMID: 16172405
- PMCID: PMC1236538
- DOI: 10.1073/pnas.0503504102
Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation
Abstract
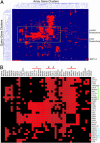
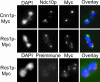
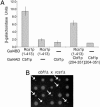
Accurate chromosome segregation requires the execution and coordination of many processes during mitosis, including DNA replication, sister chromatid cohesion, and attachment of chromosomes to spindle microtubules via the kinetochore complex. Additional pathways are likely involved because faithful chromosome segregation also requires proteins that are not physically associated with the chromosome. Using kinetochore mutants as a starting point, we have identified genes with roles in chromosome stability by performing genome-wide screens employing synthetic genetic array methodology. Two genetic approaches (a series of synthetic lethal and synthetic dosage lethal screens) isolated 211 nonessential deletion mutants that were unable to tolerate defects in kinetochore function. Although synthetic lethality and synthetic dosage lethality are thought to be based upon similar genetic principles, we found that the majority of interactions associated with these two screens were nonoverlapping. To functionally characterize genes isolated in our screens, a secondary screen was performed to assess defects in chromosome segregation. Genes identified in the secondary screen were enriched for genes with known roles in chromosome segregation. We also uncovered genes with diverse functions, such as RCS1, which encodes an iron transcription factor. RCS1 was one of a small group of genes identified in all three screens, and we used genetic and cell biological assays to confirm that it is required for chromosome stability. Our study shows that systematic genetic screens are a powerful means to discover roles for uncharacterized genes and genes with alternative functions in chromosome maintenance that may not be discovered by using proteomics approaches.
Figures

Similar articles
-
Genome-wide synthetic lethal screens identify an interaction between the nuclear envelope protein, Apq12p, and the kinetochore in Saccharomyces cerevisiae.Genetics. 2005 Oct;171(2):489-501. doi: 10.1534/genetics.105.045799. Epub 2005 Jul 5. Genetics. 2005. PMID: 15998715 Free PMC article.
-
Revealing hidden relationships among yeast genes involved in chromosome segregation using systematic synthetic lethal and synthetic dosage lethal screens.Cell Cycle. 2006 Mar;5(6):592-5. doi: 10.4161/cc.5.6.2583. Epub 2006 Mar 15. Cell Cycle. 2006. PMID: 16582600 Review.
-
Kinetochore function and chromosome segregation rely on critical residues in histones H3 and H4 in budding yeast.Genetics. 2013 Nov;195(3):795-807. doi: 10.1534/genetics.113.152082. Epub 2013 Sep 13. Genetics. 2013. PMID: 24037263 Free PMC article.
-
Suppressor analysis of a histone defect identifies a new function for the hda1 complex in chromosome segregation.Genetics. 2006 May;173(1):435-50. doi: 10.1534/genetics.105.050559. Epub 2006 Jan 16. Genetics. 2006. PMID: 16415367 Free PMC article.
-
Global synthetic-lethality analysis and yeast functional profiling.Trends Genet. 2006 Jan;22(1):56-63. doi: 10.1016/j.tig.2005.11.003. Epub 2005 Nov 23. Trends Genet. 2006. PMID: 16309778 Review.
Cited by
-
Loss of inner kinetochore genes is associated with the transition to an unconventional point centromere in budding yeast.PeerJ. 2020 Sep 29;8:e10085. doi: 10.7717/peerj.10085. eCollection 2020. PeerJ. 2020. PMID: 33062452 Free PMC article.
-
Yeast genetic interaction screens in the age of CRISPR/Cas.Curr Genet. 2019 Apr;65(2):307-327. doi: 10.1007/s00294-018-0887-8. Epub 2018 Sep 25. Curr Genet. 2019. PMID: 30255296 Free PMC article. Review.
-
Chemotherapy and signaling: How can targeted therapies supercharge cytotoxic agents?Cancer Biol Ther. 2010 Nov 1;10(9):839-53. doi: 10.4161/cbt.10.9.13738. Epub 2010 Nov 1. Cancer Biol Ther. 2010. PMID: 20935499 Free PMC article. Review.
-
Synthetic genetic array screen identifies PP2A as a therapeutic target in Mad2-overexpressing tumors.Proc Natl Acad Sci U S A. 2014 Jan 28;111(4):1628-33. doi: 10.1073/pnas.1315588111. Epub 2014 Jan 14. Proc Natl Acad Sci U S A. 2014. PMID: 24425774 Free PMC article.
-
Heterozygous screen in Saccharomyces cerevisiae identifies dosage-sensitive genes that affect chromosome stability.Genetics. 2008 Mar;178(3):1193-207. doi: 10.1534/genetics.107.084103. Epub 2008 Feb 1. Genetics. 2008. PMID: 18245329 Free PMC article.
References
-
- Uhlmann, F. (2003) Curr. Biol. 13, R104–R114. - PubMed
-
- McAinsh, A. D., Tytell, J. D. & Sorger, P. K. (2003) Annu. Rev. Cell Dev. Biol. 19, 519–539. - PubMed
-
- Cleveland, D. W., Mao, Y. & Sullivan, K. F. (2003) Cell 112, 407–421. - PubMed
-
- Rajagopalan, H. & Lengauer, C. (2004) Nature 432, 338–341. - PubMed
-
- Measday, V. & Hieter, P. (2004) Nat. Cell Biol. 6, 94–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

