Genomewide nonadditive gene regulation in Arabidopsis allotetraploids
- PMID: 16172500
- PMCID: PMC1456178
- DOI: 10.1534/genetics.105.047894
Genomewide nonadditive gene regulation in Arabidopsis allotetraploids
Abstract
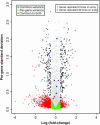
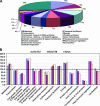
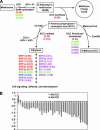
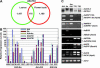
Polyploidy has occurred throughout the evolutionary history of all eukaryotes and is extremely common in plants. Reunification of the evolutionarily divergent genomes in allopolyploids creates regulatory incompatibilities that must be reconciled. Here we report genomewide gene expression analysis of Arabidopsis synthetic allotetraploids, using spotted 70-mer oligo-gene microarrays. We detected >15% transcriptome divergence between the progenitors, and 2105 and 1818 genes were highly expressed in Arabidopsis thaliana and A. arenosa, respectively. Approximately 5.2% (1362) and 5.6% (1469) genes displayed expression divergence from the midparent value (MPV) in two independently derived synthetic allotetraploids, suggesting nonadditive gene regulation following interspecific hybridization. Remarkably, the majority of nonadditively expressed genes in the allotetraploids also display expression changes between the parents, indicating that transcriptome divergence is reconciled during allopolyploid formation. Moreover, >65% of the nonadditively expressed genes in the allotetraploids are repressed, and >94% of the repressed genes in the allotetraploids match the genes that are expressed at higher levels in A. thaliana than in A. arenosa, consistent with the silencing of A. thaliana rRNA genes subjected to nucleolar dominance and with overall suppression of the A. thaliana phenotype in the synthetic allotetraploids and natural A. suecica. The nonadditive gene regulation is involved in various biological pathways, and the changes in gene expression are developmentally regulated. In contrast to the small effects of genome doubling on gene regulation in autotetraploids, the combination of two divergent genomes in allotetraploids by interspecific hybridization induces genomewide nonadditive gene regulation, providing a molecular basis for de novo variation and allopolyploid evolution.
Figures



References
-
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

