Identification of novel steroid-response elements
- PMID: 1617301
- PMCID: PMC6057364
Identification of novel steroid-response elements
Abstract
A rapid method for defining novel steroid-responsive elements has been developed. Large libraries of degenerate oligonucleotides were analyzed using a yeast-based screen to identify estrogen-responsive DNA sequences. From a library of 40,000 recombinants, seven estrogen-responsive clones were identified. When sequenced, these elements showed remarkable diversity and were different from the consensus vitellogenin A2 ERE. One surprising result was the presence of the two half sites as direct repeats in some of the clones. This implies that in vivo estrogen receptor can bind and transactivate yeast genes through response elements in which the two half sites align as direct repeats. This protocol requires no purified protein and specifically selects for functional response elements. It has a wide application in the study of any transcription factor/DNA interaction.
Figures

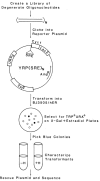

Similar articles
-
Novel estrogen response elements identified by genetic selection in yeast are differentially responsive to estrogens and antiestrogens in mammalian cells.Mol Endocrinol. 1994 Sep;8(9):1193-207. doi: 10.1210/mend.8.9.7838152. Mol Endocrinol. 1994. PMID: 7838152
-
Estrogen receptor affinity and location of consensus and imperfect estrogen response elements influence transcription activation of simplified promoters.Mol Endocrinol. 1996 Jun;10(6):694-704. doi: 10.1210/mend.10.6.8776729. Mol Endocrinol. 1996. PMID: 8776729
-
Synergism between a half-site and an imperfect estrogen-responsive element, and cooperation with COUP-TFI are required for estrogen receptor (ER) to achieve a maximal estrogen-stimulation of rainbow trout ER gene.Eur J Biochem. 1999 Jan;259(1-2):385-95. doi: 10.1046/j.1432-1327.1999.00072.x. Eur J Biochem. 1999. PMID: 9914518
-
Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action.Recent Prog Horm Res. 1997;52:141-64; discussion 164-5. Recent Prog Horm Res. 1997. PMID: 9238851 Review.
-
The role of DNA response elements as allosteric modulators of steroid receptor function.Mol Cell Endocrinol. 2005 May 31;236(1-2):1-7. doi: 10.1016/j.mce.2005.03.007. Mol Cell Endocrinol. 2005. PMID: 15876478 Review.
Cited by
-
Functional analysis of mouse Hoxa-7 in Saccharomyces cerevisiae: sequences outside the homeodomain base contact zone influence binding and activation.Mol Cell Biol. 1994 Jan;14(1):238-54. doi: 10.1128/mcb.14.1.238-254.1994. Mol Cell Biol. 1994. PMID: 8264592 Free PMC article.
-
The yeast SIN3 gene product negatively regulates the activity of the human progesterone receptor and positively regulates the activities of GAL4 and the HAP1 activator.Mol Gen Genet. 1994 Dec 15;245(6):724-33. doi: 10.1007/BF00297279. Mol Gen Genet. 1994. PMID: 7830720
-
A nuclear hormone receptor-associated protein that inhibits transactivation by the thyroid hormone and retinoic acid receptors.Proc Natl Acad Sci U S A. 1995 Oct 10;92(21):9525-9. doi: 10.1073/pnas.92.21.9525. Proc Natl Acad Sci U S A. 1995. PMID: 7568167 Free PMC article.
-
Importance of sex to pain and its amelioration; relevance of spinal estrogens and its membrane receptors.Front Neuroendocrinol. 2012 Oct;33(4):412-24. doi: 10.1016/j.yfrne.2012.09.004. Epub 2012 Oct 2. Front Neuroendocrinol. 2012. PMID: 23036438 Free PMC article. Review.
-
Estrogen receptor interaction with estrogen response elements.Nucleic Acids Res. 2001 Jul 15;29(14):2905-19. doi: 10.1093/nar/29.14.2905. Nucleic Acids Res. 2001. PMID: 11452016 Free PMC article. Review.
References
-
- Beato M. (1989), Cell 56, 335–344. - PubMed
-
- Blackwell T. K. and Weintraub H. (1990), Science 250, 1104–1110. - PubMed
-
- Bradford M. M. (1976), Anal Biochem 72, 248–254. - PubMed
-
- Carson M. A., Tsai M-J., Conneely O. M., Maxwell B. L., Clarck J. M., Dobson A. D. W., Elbrecht A., Toft D. O., Schrader W T., and O’Malley B. W. (1987), Mol Endocrinol 1, 791–801. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
