Developmental changes in the methylation status of regulatory elements in the murine alpha 1(I) collagen gene
- PMID: 1617303
- PMCID: PMC6057359
Developmental changes in the methylation status of regulatory elements in the murine alpha 1(I) collagen gene
Abstract
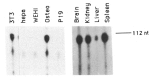
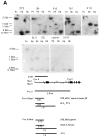
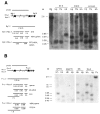
Regulatory elements contributing to the tissue-specific regulation of the murine alpha 1(I) collagen (Colla1) gene have previously been identified in its promoter region and first intron. Because several lines of evidence indicate that DNA methylation may be involved in the tissue-specific regulation of Colla1 gene expression, we have analyzed the methylation status of the 5' region of the gene by restriction analysis and a methylation-dependent PCR assay. All sites tested were unmethylated in sperm DNA. The region surrounding the start site of transcription was partially or completely methylated in undifferentiated embryonal cell lines, suggesting that it may be marked by de novo methylation during early embryonic development. In differentiated cells and adult tissues, the Colla1 promoter was completely demethylated in collagen-producing and some nonproducing cells, and partially methylated in other nonproducing cells. The first intron was unmethylated in collagen-producing as well as nonproducing cells. Only sites in the first exon showed an inverse correlation with transcriptional activity, i.e., they were unmethylated in cells that express the gene, but predominantly methylated in cells that do not. Our results indicate that the methylation status of a small area (less than 1 kb) downstream of the Colla1 promoter, but not of the promoter itself or the first intron, may be critical for transcriptional activity of the promoter, presumably through an indirect mechanism.
Figures



Similar articles
-
DNA methylation represses the murine alpha 1(I) collagen promoter by an indirect mechanism.Mol Cell Biol. 1994 Sep;14(9):5950-60. doi: 10.1128/mcb.14.9.5950-5960.1994. Mol Cell Biol. 1994. PMID: 8065328 Free PMC article.
-
Retrovirus-induced interference with collagen I gene expression in Mov13 fibroblasts is maintained in the absence of DNA methylation.Mol Cell Biol. 1991 Jan;11(1):47-54. doi: 10.1128/mcb.11.1.47-54.1991. Mol Cell Biol. 1991. PMID: 1702514 Free PMC article.
-
Variability in the upstream promoter and intron sequences of the human, mouse and chick type X collagen genes.Matrix Biol. 1996 Dec;15(6):415-22. doi: 10.1016/s0945-053x(96)90160-2. Matrix Biol. 1996. PMID: 9049979
-
The chick and human collagen alpha1(XII) gene promoter--activity of highly conserved regions around the first exon and in the first intron.Eur J Biochem. 1998 Oct 15;257(2):362-71. doi: 10.1046/j.1432-1327.1998.2570362.x. Eur J Biochem. 1998. PMID: 9826181
-
Characterization of the promoter for the alpha 1 (IV) collagen gene. DNA sequences within the first intron enhance transcription.J Biol Chem. 1988 Sep 5;263(25):12310-4. J Biol Chem. 1988. PMID: 2842328
Cited by
-
DNA methylation represses the murine alpha 1(I) collagen promoter by an indirect mechanism.Mol Cell Biol. 1994 Sep;14(9):5950-60. doi: 10.1128/mcb.14.9.5950-5960.1994. Mol Cell Biol. 1994. PMID: 8065328 Free PMC article.
-
Transforming growth factor β1 induces the expression of collagen type I by DNA methylation in cardiac fibroblasts.PLoS One. 2013;8(4):e60335. doi: 10.1371/journal.pone.0060335. Epub 2013 Apr 1. PLoS One. 2013. PMID: 23560091 Free PMC article.
-
Collagen alpha1(I) gene (COL1A1) is repressed by RFX family.J Biol Chem. 2005 Jun 3;280(22):21004-14. doi: 10.1074/jbc.M413191200. Epub 2005 Mar 23. J Biol Chem. 2005. PMID: 15788405 Free PMC article.
-
Methylation profiles of genomic DNA of mouse developmental brain detected by restriction landmark genomic scanning (RLGS) method.Nucleic Acids Res. 1993 Dec 11;21(24):5604-8. doi: 10.1093/nar/21.24.5604. Nucleic Acids Res. 1993. PMID: 8284204 Free PMC article.
-
Correct cell- and differentiation-specific expression of a murine alpha 1 (I) collagen minigene in vitro differentiating embryonal carcinoma cells.Gene Expr. 1996;6(1):35-44. Gene Expr. 1996. PMID: 8931990 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
